Key Points
Class I PITPs mediate in vivo platelet phosphoinositide signaling through both their transfer and cofactor activities.
PITPα and PITPβ have both overlapping and distinct, but nonredundant, functions.
Abstract
Platelets use signal transduction pathways facilitated by class I phosphatidylinositol transfer proteins (PITPs). The 2 mammalian class I PITPs, PITPα and PITPβ, are single PITP domain soluble proteins that are encoded by different genes and share 77% sequence identity, although their individual roles in mammalian biology remain uncharacterized. These proteins are believed to shuttle phosphatidylinositol and phosphatidylcholine between separate intracellular membrane compartments, thereby regulating phosphoinositide synthesis and second messenger formation. Previously, we observed that platelet-specific deletion of PITPα, the predominantly expressed murine PITP isoform, had no effect on hemostasis but impaired tumor metastasis formation and disrupted phosphoinositide signaling. Here, we found that mice lacking the less expressed PITPβ in their platelets exhibited a similar phenotype. However, in contrast to PITPα-null platelet lysates, which have impaired lipid transfer activity, PITPβ-null platelet lysates have essentially normal lipid transfer activity, although both isoforms contribute to phosphoinositide synthesis in vitro. Moreover, we found that platelet-specific deletion of both PITPs led to ex vivo platelet aggregation/secretion and spreading defects, impaired tail bleeding, and profound tumor dissemination. Our study also demonstrated that PITP isoforms are required to maintain endogenous phosphoinositide PtdInsP2 levels and agonist-stimulated second messenger formation. The data shown here demonstrate that the 2 isoforms are functionally overlapping and that a single isoform is able to maintain the homeostasis of platelets. However, both class I PITP isoforms contribute to phosphoinositide signaling in platelets through distinct biochemical mechanisms or different subcellular domains.
Introduction
Phosphoinositides play important roles in both signal transduction and membrane trafficking.1 The primary substrate of phosphoinositides, phosphatidylinositol (PtdIns), is produced in the endoplasmic reticulum and can be distributed by PtdIns transfer proteins (PITPs) to different cellular organelles that participate in phosphoinositide synthesis. These proteins have a hydrophobic pocket, which envelops a single phosphatidylinositol, allowing for transport through the cytoplasm to join in phosphoinositide synthesis.2 The role of PITP in mediating phosphoinositide production has been demonstrated by in vitro studies in which mammalian PITP can substitute for Sec14p, the yeast lipid transfer protein, to support phosphoinositide synthesis in yeast.3,4
PITPα and PITPβ are ubiquitous proteins found in all mammals. In COS cell lines, they are expressed in different cellular compartments, with PITPα localized at the cytosol and nucleus and PITPβ at the Golgi region.5,6 Early studies in permeabilized cells have indicated that PITP isoforms are able to support phospholipase C signaling and exocytosis by facilitating phosphoinositide synthesis.7-9 Recent studies have shown that PITP is essential for PtdIns4P-dependent recruitment of GOLPH3 to Golgi membranes, an event that is required for Golgi biogenesis during neuronal development.10,11 Taken together, the biochemical and genetic analyses performed to date indicate that PITPs are essential for most phosphoinositide-dependent cellular events.
The relative expression levels of the individual isoforms vary in different cell types.12 PITPα and PITPβ isoforms are expressed at similar levels in embryonic stem cells,13 whereas their expression varies within adult mammalian tissues. With the exception of liver, lung, and kidney, the majority of tissues express PITPα as their most abundant isoform.12 Likewise in mouse platelets, PITPα expression is sevenfold more abundant than PITPβ.14 PITPα knockout mice survive up to 2 weeks after birth but have neurological defects, gut abnormalities, and hypoglycemia.15 Mice with global deletion of PITPβ are viable and have no obvious pathological abnormality.10 The contributions of each mammalian PITP remain elusive, but some recent studies found that PITPβ was significantly upregulated in the peripheral blood of patients with human atopic dermatitis,16 and PITPβ was implicated in human epidermal growth factor signaling.17 Taken together, these findings suggest that each specific PITP isoforms may have distinct functions, as reflected in the manifestation of different human disease phenotypes.
PtdIns is an essential mediator of the signal transduction pathways in all cells, including platelets. Given that PITPs were found to facilitate PtdIns pathways in yeast, we analyzed the role of the predominant platelet isoform, PITPα, in hemostasis, thrombosis, and tumor disseminations.14 Surprisingly, platelet-specific deletion of PITPα did not disrupt ex vivo platelet function nor did it affect in vivo thrombosis and hemostasis, but it impaired signal transduction pathways that determine fibrin shroud formation around tumor cells, protecting them from immunological clearance.14,18 In this study, we sought to investigate the specific contributions of platelet PITPβ-mediated phospholipid signaling to the same biological processes. We generated mice lacking either PITPβ or both PITPα and PITPβ in their platelets and directly compared the biochemical and biologic functions of the discrete mammalian class I PITP isoforms. Here, we show that the 2 PITP isoforms function in an overlapping, yet not totally redundant, manner to support platelet homeostasis. To our knowledge, our results also provide the first in vivo evidence that PITPβ regulates platelet phosphoinositide synthesis through a mechanism that is at least partly independent of its lipid transfer activity.
Methods
Animals
Animal protocols were approved by the institutional animal care and use committees of the University of Pennsylvania. Conditional knockout (Pitpα, Pitpβ, and Pitpα/β) and transgenic (Flp and Pf4-Cre) mice with C57/BL6J genetic background were used in this study. The Pitpα conditional mouse has been described previously.14 To generate the Pitpβ conditional mouse, a 1.89kb region of the Pitpβ gene, which includes exons from 4 to 6, was flanked by insertions of loxP sites and Frt/Neo cassettes. The Neo cassette was removed by crossing with the Flp mice. Floxed mice were crossed with mice expressing Cre recombinase driven by the platelet factor-4 promoter (Pf4-Cre)19 to generate Pitpβfl/flPf4-Cre+ mice that did not express PITPβ specifically in their platelets and megakaryocytes. These mice were then crossed with Pitpαfl/flPf4-Cre+ to generate platelet-specific double-knockout Pitpαfl/fl/βfl/flPf4-Cre+ mice. The mice were maintained on standard chow. Mice of both sexes between 8 and 20 weeks of age were used for the experiments, unless otherwise noted.
Additional methods are provided in the supplemental Materials.
Results
PITPβ-null platelets are functionally normal, but the deletion of both PITPs disrupts their functions
To better understand the potential clinical significance of PITP isoforms, we analyzed the relative protein expression of the 2 class I PITP isoforms in purified human platelets. As shown (Figure 1A,B), PITPβ was approximately sixfold more abundant than PITPα. These results are in contrast with those of murine platelets, in which PITPα is more abundant than PITPβ. These data suggest that PITPβ may play a more important role than PITPα in human platelets.
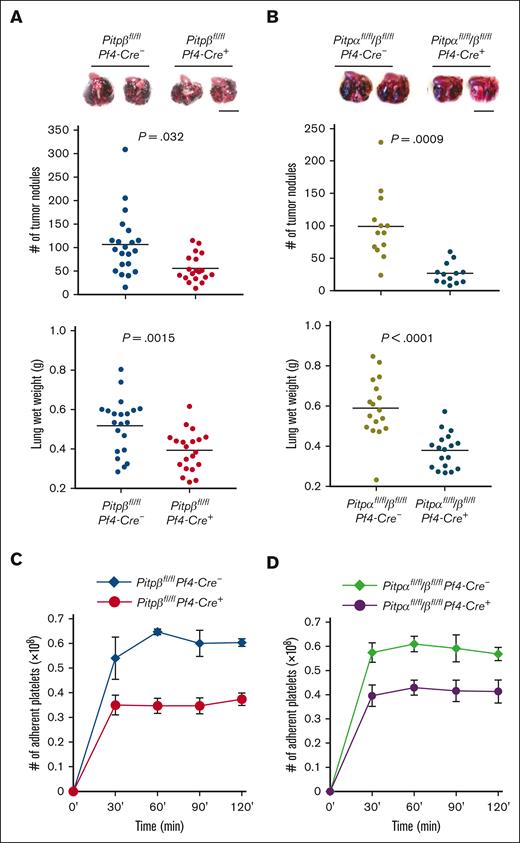
Platelet-specific loss of both PITPs impairs ex vivo platelet aggregation/secretion and spreading. (A) Western blot analysis of PITP expression in human platelets compared with their mouse counterparts. (B) Western blot–based densitometry quantification of individual PITP isoforms in human platelets. (C) Schematic representation of the conditional targeting strategy for Pitpβ. A 1.89 kb genomic DNA of PITPβ (which includes exons 4-6) was targeted by the insertion of loxP recombination sites. The Neo cassette was removed by crossing with the FRT mice before further crossing with PF4-Cre transgenic mice. (D) Western blot of platelet lysates demonstrating the specific deletion of the PITPβ isoform in Pitpβfl/flPf4-Cre+ mice. (E) Complete blood count analyses show mild thrombocytopenia in Pitpβfl/flPf4-Cre+ mice and more severe thrombocytopenia in Pitpαfl/fl/βfl/flPf4-Cre+ mice compared with their respective littermate controls (n = 6 for Pitpβfl/flPf4-Cre- mice; n = 8 for Pitpβfl/flPf4-Cre+ mice; n = 13 for Pitpαfl/fl/βfl/flPf4-Cre- mice; and n = 9 for Pitpαfl/fl/βfl/flPf4-Cre+ mice; error bars are standard deviation(s.d.); P values are shown obtained from unpaired t test). (F,G) Ex vivo analysis of platelet aggregation and dense granule secretion. PITPβ-null platelets aggregate normally, but dense granule secretion was impaired in response to low-dose thrombin (0.05 U/mL) and collagen (10 μg/mL), as measured by adenosine triphosphate release (F). Deleting both PITP isoforms increased the severity of aggregation and secretion defects (G). Adenosine triphosphate secretion traces start at 100% and trend downward, and aggregation traces start at 0% and trend upward. Traces are representative of 5 separate experiments per condition. (H) Spreading of PITPβ-null and PITPα/β-null platelets on fibrinogen after stimulation with thrombin (0.025 U/mL) revealed that PITPα/β-null platelets had a spreading defect, whereas PITPβ-null platelets spread normally. (I) PITPα/β-null platelet spreading was quantified as the total cumulative area of platelets per field (n = 3 per group). (J) The number of adherent PITPα/β-null platelets was quantified as the average number per field under a 100× microscope. (n = 5 per group; error bars represent s.d.; P values are shown, unpaired t test).
Platelet-specific loss of both PITPs impairs ex vivo platelet aggregation/secretion and spreading. (A) Western blot analysis of PITP expression in human platelets compared with their mouse counterparts. (B) Western blot–based densitometry quantification of individual PITP isoforms in human platelets. (C) Schematic representation of the conditional targeting strategy for Pitpβ. A 1.89 kb genomic DNA of PITPβ (which includes exons 4-6) was targeted by the insertion of loxP recombination sites. The Neo cassette was removed by crossing with the FRT mice before further crossing with PF4-Cre transgenic mice. (D) Western blot of platelet lysates demonstrating the specific deletion of the PITPβ isoform in Pitpβfl/flPf4-Cre+ mice. (E) Complete blood count analyses show mild thrombocytopenia in Pitpβfl/flPf4-Cre+ mice and more severe thrombocytopenia in Pitpαfl/fl/βfl/flPf4-Cre+ mice compared with their respective littermate controls (n = 6 for Pitpβfl/flPf4-Cre- mice; n = 8 for Pitpβfl/flPf4-Cre+ mice; n = 13 for Pitpαfl/fl/βfl/flPf4-Cre- mice; and n = 9 for Pitpαfl/fl/βfl/flPf4-Cre+ mice; error bars are standard deviation(s.d.); P values are shown obtained from unpaired t test). (F,G) Ex vivo analysis of platelet aggregation and dense granule secretion. PITPβ-null platelets aggregate normally, but dense granule secretion was impaired in response to low-dose thrombin (0.05 U/mL) and collagen (10 μg/mL), as measured by adenosine triphosphate release (F). Deleting both PITP isoforms increased the severity of aggregation and secretion defects (G). Adenosine triphosphate secretion traces start at 100% and trend downward, and aggregation traces start at 0% and trend upward. Traces are representative of 5 separate experiments per condition. (H) Spreading of PITPβ-null and PITPα/β-null platelets on fibrinogen after stimulation with thrombin (0.025 U/mL) revealed that PITPα/β-null platelets had a spreading defect, whereas PITPβ-null platelets spread normally. (I) PITPα/β-null platelet spreading was quantified as the total cumulative area of platelets per field (n = 3 per group). (J) The number of adherent PITPα/β-null platelets was quantified as the average number per field under a 100× microscope. (n = 5 per group; error bars represent s.d.; P values are shown, unpaired t test).
In order to understand the role of PITPβ in platelet activation, we generated mice that specifically lacked PITPβ in their platelets and megakaryocytes by crossing Pitpβfl/fl mice (Figure 1C) with Pf4-Cre+ mice.19 Mice lacking PITPβ only in their platelets and megakaryocytes were viable and produced platelets. Western blot analysis of platelet lysates probed with an anti-PITPβ–specific antibody indicated the successful deletion of the protein (Figure 1D). In the PITPβ-null platelets, we did not observe a compensatory increase of PITPα. Furthermore, by pairing Pitpαfl/flPf4-Cre+ mice with Pitpβfl/flPf4-Cre+ mice, we generated Pitpαfl/fl/βfl/flPf4-Cre+ mice that lacked both PITP isoforms in their platelets and megakaryocytes. Mice with the deletion of both isoforms (Pitpαfl/fl/βfl/flPf4-Cre+) were grossly healthy, with no apparent abnormalities in body weight, organ morphology, and survival. We also did not observe any spontaneous thrombosis or hemorrhage. However, Pitpβfl/flPf4-Cre+ mice had minimal thrombocytopenia (85% healthy), whereas Pitpαfl/fl/βfl/flPf4-Cre+ mice had even more pronounced thrombocytopenia (60% healthy; Figure 1E). The complete blood cell count in our previous study demonstrated that Pitpαfl/fl/βfl/flPf4-Cre+ mice had a slightly reduced white blood cell count (24%),20 but Pitpβfl/flPf4-Cre+ mice had healthy blood cell counts (supplemental Table 1).
Given that PITP is essential for phosphoinositide production in yeast and that phosphoinositide signaling is critical in platelet physiology, we examined whether the deletion of PITPβ or both PITP isoforms in platelets affects ex vivo activation. Similar to PITPα-null platelets,14 PITPβ-null platelets aggregated normally after stimulation with low doses of thrombin or collagen. However, PITPβ-null platelets had impaired adenosine triphosphate secretion compared with controls (Figure 1F). When both isoforms were deleted, platelets showed severe ex vivo aggregation and secretion defects (Figure 1G). Interestingly, levels of serotonin, which is stored in dense granules, were normal in platelets lacking either PITPβ or both PITP (supplemental Figure 3), suggesting that these platelets may have secretion defects rather than biogenesis or loading defects. Finally, activated PITPβ-null platelets spread normally on fibrinogen, whereas PITPα/β-null platelets were impaired (Figure 1H-J). These findings demonstrate that both isoforms play roles in regulating platelet function and that either isoform is able to maintain healthy platelet function.
Mice lacking platelet PITPs have no defects in laser-induced in vivo thrombosis but have prolonged tail bleeding times
Next, we assessed whether ex vivo platelet activation defects found in Pitpαfl/fl/βfl/flPf4-Cre+ mice (PITPα/β-null) disrupted in vivo thrombosis by using 2 models: a laser injury-induced arteriolar thrombosis model and tail bleeding time assay. After initiating laser-induced vascular injury of the cremaster arterioles, we visualized platelet accumulation (CD41) and α-granule secretion (P-selectin) in real time. Mice with PITPα/β-null platelets had no discernable changes in platelet accumulation (Figure 2A,B), and exocytosis of α-granules was normal (Figure 2C) despite the lack of granule contents, as demonstrated by electron microscopy previously in MKs.20 Next, we analyzed hemostasis using an independent in vivo model, the tail bleeding time assay. Bleeding times were normal in Pitpβfl/flPf4-Cre+ mice (Figure 2D), but there was a statistically significant delay in Pitpαf/f/βfl/flPf4-Cre+ mice compared with the littermate controls (150 seconds vs 35 seconds; P < .05; Figure 2E), revealing a contribution of PITP-dependent signaling pathways to hemostasis, which is different from that shown by the laser injury model.
Mice lacking platelet PITPs have prolonged tail bleeding time, but no defects in laser-induced in vivo thrombosis. The laser-induced injury model demonstrates normal in vivo thrombosis and platelet secretion in the Pitpαfl/fl/βfl/flPf4-Cre+ mice. (A) Representative images show platelet accumulation (CD41, red) and P-selectin exposure (green [overlay of red/green is yellow]) 3 minutes after laser-induced injury to the cremaster arterioles. Images are binary representations of 2D confocal fluorescence images overlaid on the bright-field. White arrows indicate the direction of flow; scale bar, 10 μm. (B) Graphs of the CD41+ area over time (left, mean ± standard error of the mean), median CD41+ area over time (middle), and CD41 peak area (right, lines represent median ± interquartile range). (C) Graphs of the P-selectin–positive area over time (left, mean ± standard error of the mean), the median P-selectin–positive area over time (middle), and P-selectin peak area (right, lines are median ± interquartile range). n = 20 thrombi in 4 wild-type mice and n = 29 thrombi in 4 Pitpαfl/fl/βfl/flPf4-Cre+ mice. Statistical analysis were performed using a two-tailed Mann-Whitney test. (D,E) Tail bleeding times in mice lacking PITPβ (D) or both PITP isoforms compared with their littermate controls (E). Tail bleeding was normal in mice with platelets lacking PITPβ (NS, Mann-Whitney test; n = 52 for Pitpβfl/flPf4-Cre- mice and n = 58 for Pitpβfl/flPf4-Cre+ mice). When both PITP isoforms were deleted in platelets, there was a mild increase in bleeding time (P = .0013 using two-tailed Mann-Whitney test; n = 57 for Pitpαfl/fl/βfl/flPf4-Cre- mice and n = 54 for Pitpαfl/fl/βfl/flPf4-Cre+ mice).
Mice lacking platelet PITPs have prolonged tail bleeding time, but no defects in laser-induced in vivo thrombosis. The laser-induced injury model demonstrates normal in vivo thrombosis and platelet secretion in the Pitpαfl/fl/βfl/flPf4-Cre+ mice. (A) Representative images show platelet accumulation (CD41, red) and P-selectin exposure (green [overlay of red/green is yellow]) 3 minutes after laser-induced injury to the cremaster arterioles. Images are binary representations of 2D confocal fluorescence images overlaid on the bright-field. White arrows indicate the direction of flow; scale bar, 10 μm. (B) Graphs of the CD41+ area over time (left, mean ± standard error of the mean), median CD41+ area over time (middle), and CD41 peak area (right, lines represent median ± interquartile range). (C) Graphs of the P-selectin–positive area over time (left, mean ± standard error of the mean), the median P-selectin–positive area over time (middle), and P-selectin peak area (right, lines are median ± interquartile range). n = 20 thrombi in 4 wild-type mice and n = 29 thrombi in 4 Pitpαfl/fl/βfl/flPf4-Cre+ mice. Statistical analysis were performed using a two-tailed Mann-Whitney test. (D,E) Tail bleeding times in mice lacking PITPβ (D) or both PITP isoforms compared with their littermate controls (E). Tail bleeding was normal in mice with platelets lacking PITPβ (NS, Mann-Whitney test; n = 52 for Pitpβfl/flPf4-Cre- mice and n = 58 for Pitpβfl/flPf4-Cre+ mice). When both PITP isoforms were deleted in platelets, there was a mild increase in bleeding time (P = .0013 using two-tailed Mann-Whitney test; n = 57 for Pitpαfl/fl/βfl/flPf4-Cre- mice and n = 54 for Pitpαfl/fl/βfl/flPf4-Cre+ mice).
Loss of PITPs impairs tumor metastasis and platelet thrombin generation
In addition to hemostasis, platelets have been implicated in other processes, such as the dissemination of tumor metastases through effects of the coagulation system.21,22 We have previously demonstrated that PITPα-mediated phosphoinositide metabolism within platelets enables tumor cells to escape immune surveillance that requires platelet thrombin generation and fibrin formation.14 To understand whether PITPβ also contributes to this process, we injected B16F10 melanoma cells IV and analyzed the tumor metastases in the lungs of mice lacking either PITPβ or both PITPα and PITPβ in their platelets.
We found that mice lacking platelet PITPβ had impaired tumor metastasis formation (Figure 3A), similar to mice lacking platelet PITPα.14 In addition, mice lacking both platelet PITP showed an even greater reduction in tumor metastasis than mice lacking a single isoform (Figure 3B). We previously demonstrated that the ability of platelets to support metastasis formation correlates with their ability to adhere to tumor cells.14 Therefore, we analyzed the adhesion of PITPβ-null platelets to tumor cells. We found that PITPβ deletion contributed to the loss of platelet-tumor adhesion, comparable with PITPα deletion (Figure 3C), and the loss of both PITP isoforms did not further compound the adhesion defect (Figure 3D), revealing that adhesion is not the only mechanism driving metastasis formation.
Platelets lacking PITPβ or both PITP isoforms are less susceptible to tumor metastasis. (A,B) Lungs harvested from Pitpβfl/flPf4-Cre- and Pitpβfl/flPf4-Cre+ mice (A) or Pitpαfl/fl/βfl/flPf4-Cre- and Pitpαfl/fl/βfl/flPf4-Cre+ mice (B) 2 weeks after tail vein injection with B16F10 melanoma cells demonstrated that loss of PITP impairs metastasis. Representative lungs are shown at 2 weeks after tumor cell injection (top); the number of tumor nodules on the lung surface 2 weeks after tumor cell injection (middle); and wet lung weights 3 weeks after tumor injection (bottom). For tumor nodule counting, n = 21 lungs for Pitpβfl/flPf4-Cre- mice, n = 19 for Pitpβfl/flPf4-Cre+ mice, and n = 13 for both Pitpαfl/fl/βfl/flPf4-Cre- mice and Pitpαfl/fl/βfl/flPf4-Cre+ mice. For lung weight, n = 21 lungs for Pitpβfl/flPf4-Cre- mice, n = 20 for Pitpβfl/flPf4-Cre+ mice, n = 17 for Pitpαfl/fl/βfl/flPf4-Cre- mice, and n = 18 for Pitpαfl/fl/βfl/flPf4-Cre+ mice. Statistical analysis was performed using an unpaired t test. Black scale bars represent 10 mm. (C,D) Ex vivo adhesion of PITPβ-null (C) or PITPα/β-null (D) platelets to a tissue-cultured tumor cell monolayer was impaired compared with wild-type controls. Error bars represent s.d.; n = 3 for each genotype.
Platelets lacking PITPβ or both PITP isoforms are less susceptible to tumor metastasis. (A,B) Lungs harvested from Pitpβfl/flPf4-Cre- and Pitpβfl/flPf4-Cre+ mice (A) or Pitpαfl/fl/βfl/flPf4-Cre- and Pitpαfl/fl/βfl/flPf4-Cre+ mice (B) 2 weeks after tail vein injection with B16F10 melanoma cells demonstrated that loss of PITP impairs metastasis. Representative lungs are shown at 2 weeks after tumor cell injection (top); the number of tumor nodules on the lung surface 2 weeks after tumor cell injection (middle); and wet lung weights 3 weeks after tumor injection (bottom). For tumor nodule counting, n = 21 lungs for Pitpβfl/flPf4-Cre- mice, n = 19 for Pitpβfl/flPf4-Cre+ mice, and n = 13 for both Pitpαfl/fl/βfl/flPf4-Cre- mice and Pitpαfl/fl/βfl/flPf4-Cre+ mice. For lung weight, n = 21 lungs for Pitpβfl/flPf4-Cre- mice, n = 20 for Pitpβfl/flPf4-Cre+ mice, n = 17 for Pitpαfl/fl/βfl/flPf4-Cre- mice, and n = 18 for Pitpαfl/fl/βfl/flPf4-Cre+ mice. Statistical analysis was performed using an unpaired t test. Black scale bars represent 10 mm. (C,D) Ex vivo adhesion of PITPβ-null (C) or PITPα/β-null (D) platelets to a tissue-cultured tumor cell monolayer was impaired compared with wild-type controls. Error bars represent s.d.; n = 3 for each genotype.
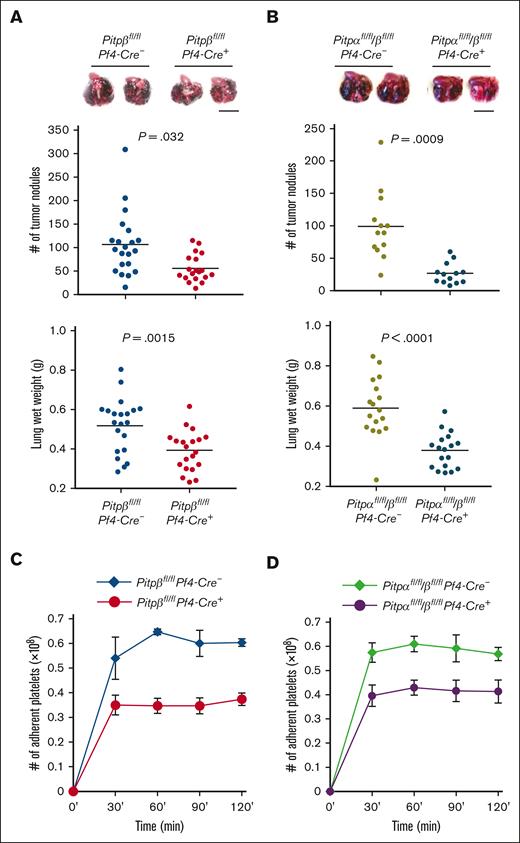
Our previous study demonstrated that tumor cells can escape host immune surveillance by inducing thrombin generation on its surface, causing platelets to agglutinate and to shroud the tumor cells from immune cells.14 We observed that, like PITPα, platelet PITPβ facilitates tumor-induced thrombin generation, and in the absence of both PITP isoforms, thrombin generation in platelet-rich plasma was essentially eliminated (Figure 4A,B). Because the tissue factor (TF) on the tumor surface is likely to trigger thrombin production in platelets, we tested the ability of PITP-null platelets to generate thrombin when stimulated with recombinant TF alone. As predicted, TF-induced thrombin generation was markedly impaired in PITPβ-null platelets and was, essentially, eliminated in platelets lacking both the PITP isoforms (Figure 4C,D). In both cases, thrombin generation in platelet-poor plasma was identical in PITP-null and wild-type cells after stimulation with either tumor cells (Figure 4E) or TF (Figure 4F), confirming that PITP-mediated thrombin generation is platelet-dependent. Together, these results demonstrate that both PITP isoforms contribute to tumor dissemination via a pathway that is dependent on platelet PITP-mediated thrombin generation.
Loss of PITP in platelets impairs thrombin generation and Annexin V binding. (A-D) Representative kinetics of thrombin generation induced by BF610 tumor cells (A,B) or TF (C,D) in platelet-rich plasma (PRP) from Pitpβfl/flPf4-Cre- mice (wild-type control, navy trace), Pitpβfl/flPf4-Cre+ mice (PITPβ-null, red trace), and Pitpαfl/fl/βfl/flPf4-Cre+ mice (PITPα/β-null, purple trace). Endogenous thrombin potential is shown as the mean value of total thrombin induced by B16F10 tumor cells (B) or by TF (D) over 90 minutes of reaction time in PRP containing PITPβ-null platelets (red bars), PITPα/β-null platelets (purple bars), and their wild-type controls (navy bars). (E,F) Platelet-poor plasma (PPP) was used as a control to demonstrate the platelet-intrinsic nature of thrombin generation upon stimulation with B16F10 tumor cells (E) or TF (F). n = 3 mice per group. Statistical analysis was performed using an unpaired t test. Error bars are s.d. (G) Platelets lacking PITPβ and both PITP isoforms have an impaired ability to bind Annexin V after activation by the combination of 5 μg/mL collagen and 0.05 U/mL thrombin. The mean values are averaged from 4 independent experiments. Data were analyzed using unpaired t test. Error bars are s.d.
Loss of PITP in platelets impairs thrombin generation and Annexin V binding. (A-D) Representative kinetics of thrombin generation induced by BF610 tumor cells (A,B) or TF (C,D) in platelet-rich plasma (PRP) from Pitpβfl/flPf4-Cre- mice (wild-type control, navy trace), Pitpβfl/flPf4-Cre+ mice (PITPβ-null, red trace), and Pitpαfl/fl/βfl/flPf4-Cre+ mice (PITPα/β-null, purple trace). Endogenous thrombin potential is shown as the mean value of total thrombin induced by B16F10 tumor cells (B) or by TF (D) over 90 minutes of reaction time in PRP containing PITPβ-null platelets (red bars), PITPα/β-null platelets (purple bars), and their wild-type controls (navy bars). (E,F) Platelet-poor plasma (PPP) was used as a control to demonstrate the platelet-intrinsic nature of thrombin generation upon stimulation with B16F10 tumor cells (E) or TF (F). n = 3 mice per group. Statistical analysis was performed using an unpaired t test. Error bars are s.d. (G) Platelets lacking PITPβ and both PITP isoforms have an impaired ability to bind Annexin V after activation by the combination of 5 μg/mL collagen and 0.05 U/mL thrombin. The mean values are averaged from 4 independent experiments. Data were analyzed using unpaired t test. Error bars are s.d.
Thrombin generation depends on the flipping of phosphatidylserine (PS) from the inner to the outer leaflet of the platelet cell membrane. To determine whether platelets lacking PITPβ or both PITP isoforms have impaired PS exposure, we analyzed Annexin V–binding to detect PS on the surface of platelets. In resting platelets, PS exposure was minimal and unaffected by the absence of either of the PITP isoforms (Figure 4G). However, PS exposure was significantly reduced in activated platelets lacking PITPβ, similar to that in our previous study on PITPα,14 and was further reduced when both isoforms were deleted. The ability of PITPs to support the flipping of PS on the cell membrane likely accounts for the mechanism by which PITP enables tumor-mediated thrombin generation.
PITPβ is required to maintain endogenous phospholipid homeostasis
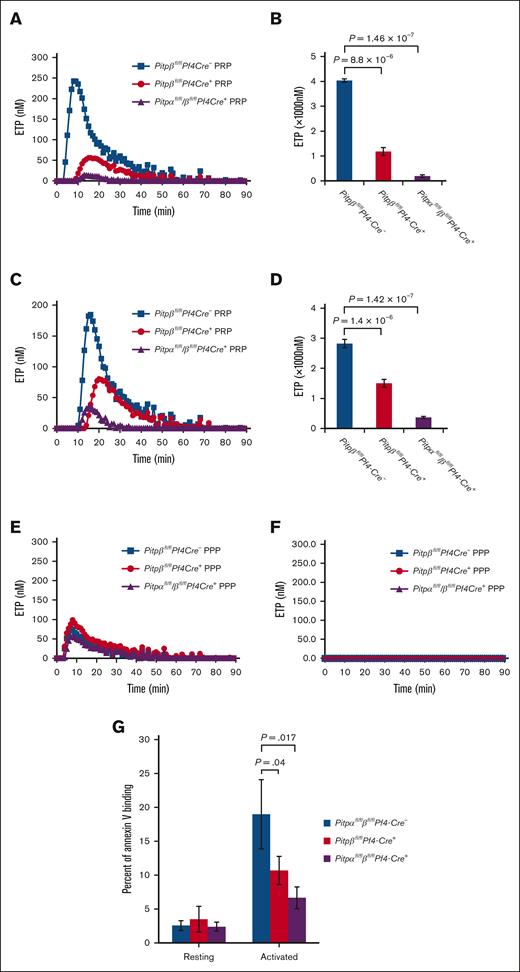
PITPs are known for their ability to transport PtdIns from the endoplasmic reticulum to different subcellular compartments to facilitate intracellular phospholipid signaling. We analyzed the relative contributions of either isoform to the synthesis of platelet phosphoinositide PtdInsP and PtdInsP2 via mass spectrometry. The most abundant phosphoinositide was distributed in a fraction of C38:4, and the least abundant phosphoinositide was found in a fraction of C36:2 (supplemental Figure 4A). Under both resting and thrombin-activated conditions, platelets lacking either PITPα or PITPβ showed no significant reduction in total endogenous PtdInsP, including PtdIns(3)P, PtdIns(4)P, and PtdIns(5)P. However, platelets lacking both isoforms had markedly reduced amounts of endogenous PtdInsP, and thrombin activation failed to induce additional PtdInsP production (Figure 5A; supplemental Figure 4A). The data shown here suggest that either isoform alone can maintain sufficient PtdInsP required for normal platelet function. Surprisingly, PITPβ can compensate for the loss of PITPα to maintain PtdInsP levels, even though there is relatively little PITPβ in the platelets.
Mass spectrometry analysis of endogenous phosphoinositide and IP3 production in murine platelets. (A) Endogenous levels of PtdInsP(PIP) in platelets lacking a single isoform of PITPα, PITPβ, or both. The total PIP response include PtdIns(3)P, PtdIns(4)P, and PtdIns(5)P in all fractions, including C38:4, C38:3, and C36:2. (B) PtdInsP2 production in platelets with the deletion of either a single isoform or both PITPs. The total PtdInsP2 response include PtdIns(3,4)P2, PtdIns(3,5)P2, and PtdIns(4,5)P2 in all the fractions. The assay was repeated 3 times for each group. The data represent endogenous phosphoinositide levels in 5 × 106 cells and normalized by adding known amounts of phosphoinositide as an internal control (mean ± s.d.). IP3 production was impaired in thrombin-stimulated (1 U/mL for 1 minute) PITPβ-null (C, red trace) platelets and PITPα/β-null (D, teal trace) platelets compared with wild-type littermate controls.
Mass spectrometry analysis of endogenous phosphoinositide and IP3 production in murine platelets. (A) Endogenous levels of PtdInsP(PIP) in platelets lacking a single isoform of PITPα, PITPβ, or both. The total PIP response include PtdIns(3)P, PtdIns(4)P, and PtdIns(5)P in all fractions, including C38:4, C38:3, and C36:2. (B) PtdInsP2 production in platelets with the deletion of either a single isoform or both PITPs. The total PtdInsP2 response include PtdIns(3,4)P2, PtdIns(3,5)P2, and PtdIns(4,5)P2 in all the fractions. The assay was repeated 3 times for each group. The data represent endogenous phosphoinositide levels in 5 × 106 cells and normalized by adding known amounts of phosphoinositide as an internal control (mean ± s.d.). IP3 production was impaired in thrombin-stimulated (1 U/mL for 1 minute) PITPβ-null (C, red trace) platelets and PITPα/β-null (D, teal trace) platelets compared with wild-type littermate controls.
We also analyzed the total PtdInsP2 levels [PtdIns(3,4)P2, PtdIns(3,5)P2, and PtdIns(4,5)P2] in platelets derived from all fractions (C38:4, C38:3, and C36:2). In contrast to PtdInsP production, we found that platelets lacking either a single isoform or both PITPs had only minor and PI3 kinase–independent reduction of endogenous PtdInsP2 analyzed at resting conditions and after thrombin stimulation (Figure 5B; supplemental Figure 4A-B).
In our studies of platelets lacking PIP5KI, we observed that phospholipase C generates the second messenger inositol trisphosphate (IP3), which is derived from pools of PtdIns(4,5)P2 within specific microdomains.23 We hypothesized that the loss of individual PITP isoforms could impair the synthesis of discrete pools of PtdIns(4,5)P2 required for second messenger formation without markedly affecting the levels of total cellular PtdIns(4,5)P2. To test this hypothesis, we analyzed IP3 production following thrombin activation in platelets lacking either PITPβ or both isoforms.
Although activation caused a rapid increase in IP3 formation in wild-type platelets, IP3 production in PITPβ-null platelets was reduced by up to 50% following activation (Figure 5C), similar to the previously reported result found in PITPα-null platelets.14 Further, platelets lacking both isoforms had PLC activity independent, essentially no IP3 production following stimulation (Figure 5D; supplemental Figure 4C). These data indicate that although PITP isoforms are not critical for the synthesis of total cellular PtdInsP2, they are essential for the synthesis of discrete pools of PtdIns(4,5)P2. Because PITPα is more abundant than PITPβ, these data also demonstrate the unexpectedly large contribution of PITPβ to the discrete pools of PtdIns(4,5)P2 required for IP3 production.
PITPβ is a cofactor for phosphoinositide synthesis
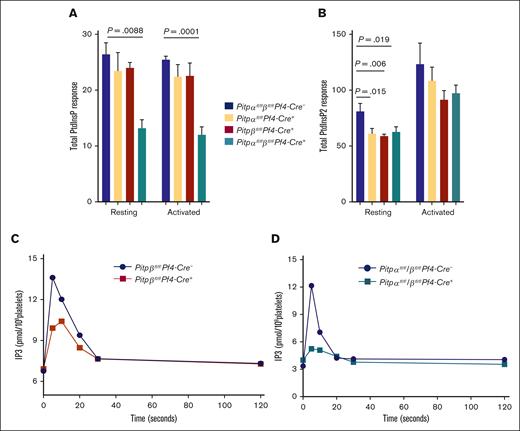
Because the primary function of PITPs has been discovered by their ability to transfer PtdIns, we compared the in vitro transfer activity of tritiated phosphatidylinositol from microsomes to liposomes in the presence of lysates of PITPα-null and PITPβ-null platelets.24 As predicted, when both PITP isoforms were deleted in platelets, the PtdIns transfer activity was completely ablated (Figure 6A). Interestingly, PITPα-null platelet lysates showed more than 90% reduction in transfer activity compared with the lysates from the control platelets. However, PITPβ-null platelet lysates showed almost no reduction in PtdIns transfer (Figure 6A). These data demonstrate that PITPα is responsible for all the essentially PtdIns transfer activities within platelets. The difference in the transfer activity mediated by PITPα and PITPβ could be due to the difference in the levels of PITP protein expression within platelet.14 Importantly, these data indicate that PITPβ does not support PtdIns transfer activity in platelets in a significant way, and thus, its effect on PtdInsP2 production is mediated through another mechanism.
PITPβ does not have transfer activity but has cofactor activity. (A) In vitro [3H]-labeled PtdIns transfer activity from microsomes (permeabilized HL60 cells) to liposomes (PC:PI :: 98:2) is mediated by platelet PITPα, but not PITPβ. (B.C) Lipid kinase assays were performed to determine the effects of platelet PITPα (left) and PITPβ (right) on PtdInsP synthesis in vitro, before and after thrombin stimulation (3 minutes: time of thrombin stimulation [1 U/mL]). This assay, which does not require transfer activity, demonstrated that both PITP isoforms are required for phospholipid kinases to generate phosphoinositides. Phosphoinositide production in PITP-null platelets was restored by the addition of rPITPα and rPITPβ. ∗∗P < .01.
PITPβ does not have transfer activity but has cofactor activity. (A) In vitro [3H]-labeled PtdIns transfer activity from microsomes (permeabilized HL60 cells) to liposomes (PC:PI :: 98:2) is mediated by platelet PITPα, but not PITPβ. (B.C) Lipid kinase assays were performed to determine the effects of platelet PITPα (left) and PITPβ (right) on PtdInsP synthesis in vitro, before and after thrombin stimulation (3 minutes: time of thrombin stimulation [1 U/mL]). This assay, which does not require transfer activity, demonstrated that both PITP isoforms are required for phospholipid kinases to generate phosphoinositides. Phosphoinositide production in PITP-null platelets was restored by the addition of rPITPα and rPITPβ. ∗∗P < .01.
In addition to phospholipid transfer, recent studies have demonstrated that PITPs directly interact with lipid kinases to enhance their kinetics.1,25,26 Thus, we analyzed the potential impact of each individual isoform on platelet lipid kinase activity in mediating PtdInsP synthesis in vitro. We used a cell-free system with excess lipid substrate to assess the cofactor capability of PITP to enhance lipid kinase function. Because lipid availability was not rate limiting and cellular compartments were eliminated, this assay analyzed each individual PITP isoform for their so-called “nanoreactor” ability to assist phosphoinositide kinases through a lipid transfer–independent mechanism.25 The data show that PITPα-null platelet lysate have reduced PtdInsP production after thrombin activation. Augmentation with either recombinant PITPα (rPITPα) or rPITPβ completely restored PtdInsP production (Figure 6B). Conversely, lysates from PITPβ-null platelets also showed reduced PtdInsP production, similar to PITPα-null platelet lysate, and augmentation with rPITPα or rPITPβ restored the ability of PITPβ-null platelet lysates to generate PtdInsP (Figure 6C), indicating that each isoform can compensate for the loss of the deleted isoform in mediating PtdInsP synthesis in vitro. Taken together, to our knowledge, these results provide the first evidence that both platelet PITP isoforms function as cofactor in phosphoinositide synthesis.
PITP isoforms distributed differentially within platelets
Given that PITPα is the predominant isoform involved in lipid transfer in platelets, both isoforms are interchangeable in their ability to provide a cofactor activity to lipid kinases. We hypothesized that these isoforms are differentially distributed within platelets and contribute to phosphoinositide synthesis in different subcellular compartments. To address this hypothesis, we first fractionated wild-type platelets into cytosolic, membrane, and cytoskeletal fractions and immunoblotted them for PITPα and PITPβ. We found that PITPα was overwhelmingly localized in the cytosol (89.5%), with very little found in the membrane (4.8%) and cytoskeletal (5.7%) fractions. In contrast, PITPβ localization was more dispersed in the cytosolic (62.8%), membrane (13.2%), and cytoskeletal fractions (24%). Finally, thrombin activation did not change the distribution of any isoforms (Figure 7A,B). Next, we investigated the cellular distribution of PITPα in PITPβ-null platelets and, conversely, the PITPβ distributions in PITPα-null platelets. PITPα distribution in both resting and activated PITPβ-null platelets was shifted away from the cytosol to the membrane (14.7%) and cytoskeleton (25.3%) fractions compared with the wild-type (Figure 7C,D). Furthermore, the PITPβ distribution in PITPα-null platelets shifted away from the membrane and cytoskeleton to the point that it was undetectable, and toward the cytosol (Figure 7E). Thus, in both cases, the distribution of the compensating isoform shifted toward the distribution profile of the missing isoform, suggesting spatial compensation of the missing isoform. We further examined human platelets, and the spatial distribution of the isoforms was quite different from that of mouse platelets, as the majority of PITPα was found in the membrane (up to 60%), whereas the cytoskeleton contained the least amount of protein (Figure 7F,G). However, PITPβ was distributed predominantly in the cytosol and cytoskeleton and was undetectable in the membrane fraction (Figure 7F,G). These data demonstrate that distinct PITP isoforms diverge in their spatial localization within human platelets, which could explain their differing roles in platelet signal transduction.
Fractioned distribution of PITPα and PITPβ within the platelets. (A) Representative immunoblot and (B) densitometry quantification of wild-type platelets indicates that PITPα is mostly distributed in the cytosol, whereas PITPβ has a disproportionate amount of total protein in the membrane and cytoskeleton (n = 3 separate experiments) in both resting and thrombin-activated platelets (1 U/mL for 1 minute). (C) Representative immunoblot of the fractioned distribution of PITPα in platelets lacking PITPβ. (D) Densitometry quantification of PITPα fractioned distribution in platelets lacking PITPβ. (E) Distribution of PITPβ in platelets lacking PITPα (bottom). (F) Fractioned distribution of PITPα and PITPβ in resting (left) and thrombin-activated (right) human platelets. (G) Densitometry quantification of PITPα and PITPβ in different cellular fractions of resting human platelets. All densitometry data from 3 separate experiments were summed and the percentage relative to the total single PITP isoform was plotted for each fraction. (H) In this model, PITPα serves the traditional role of transferring PtdIns from 1 subcellular compartment to another, such as the plasma membrane, and PITPβ, in turn, serves as a cofactor for PI kinase–mediated PIP synthesis.
Fractioned distribution of PITPα and PITPβ within the platelets. (A) Representative immunoblot and (B) densitometry quantification of wild-type platelets indicates that PITPα is mostly distributed in the cytosol, whereas PITPβ has a disproportionate amount of total protein in the membrane and cytoskeleton (n = 3 separate experiments) in both resting and thrombin-activated platelets (1 U/mL for 1 minute). (C) Representative immunoblot of the fractioned distribution of PITPα in platelets lacking PITPβ. (D) Densitometry quantification of PITPα fractioned distribution in platelets lacking PITPβ. (E) Distribution of PITPβ in platelets lacking PITPα (bottom). (F) Fractioned distribution of PITPα and PITPβ in resting (left) and thrombin-activated (right) human platelets. (G) Densitometry quantification of PITPα and PITPβ in different cellular fractions of resting human platelets. All densitometry data from 3 separate experiments were summed and the percentage relative to the total single PITP isoform was plotted for each fraction. (H) In this model, PITPα serves the traditional role of transferring PtdIns from 1 subcellular compartment to another, such as the plasma membrane, and PITPβ, in turn, serves as a cofactor for PI kinase–mediated PIP synthesis.
Discussion
In this study, we sought to determine how PITPβ contributes to intracellular platelet signaling, platelet activation, hemostasis, and nonhemostatic tumor metastasis formation. We were also interested in comparing the results with those of a previous PITPα study to determine any redundancies or differences in function. Our major findings are as follows: (1) PITPβ is the most abundant isoform in human platelets; (2) Mice lacking PITPβ or both PITP isoforms in their megakaryocytes and platelets can survive normally but develop thrombocytopenia; (3) Platelets have a severe aggregation and dense granule defect when both PITP isoforms are deleted; (4) Deletion of the PITPβ isoform had effects comparable with those of PITPα on the levels of endogenous PtdInsP, PtdInsP2, and agonist-induced second messenger production, indicating that PITPβ is as important as PITPα in platelet signal transduction; (5) Like PITPα, PITPβ contributes to a process that enables tumors to escape innate immunity; (6) PITPα contributes to the majority of PtdIns transfer activity in murine platelets; (7) Both PITP isoforms serve as cofactors for lipid kinases, and either isoform can functionally compensate for the loss of another (Table 1); and (8) PITP isoforms likely exert distinct contributions to platelet biology by regulating spatially distinct pools or pathways of phosphoinositide, as demonstrated by their differential intracellular compartmentalization.
Unlike deletions in other tissues, such as PITPα in neurons or PITPβ in embryonic stem cells,13 the deletion of PITP in murine platelets did not cause a severe phenotype. Dense granule secretion and platelet spreading in PITPβ-null platelets were partially reduced. However, when both isoforms were deleted, platelet aggregation, secretion, and spreading were significantly impaired with a concomitant marked reduction in phosphoinositide production. We found that each PITP isoform can compensate for the loss of the other isoform, demonstrating that these isoforms have overlapping but complementary roles in facilitating phospholipid signaling, which drives platelet shape change and granule secretion. Strikingly, significant in vivo impairment was absent in the laser injury-induced thrombosis model, even when both isoforms were deleted in the platelets. This suggests that sufficient platelet phosphoinositide was produced, despite PITP deletion, to recruit platelets for normal hemostasis, perhaps facilitated by other PITP classes or mechanisms independent of PITP. Studies have found that PITPα associated with in vivo pathogenesis is largely independent of the levels of protein expression, and only 10% of normal expression can support the minimum homeostasis requirements.27 However, in other in vivo assays, such as tail bleeding time and tumor metastasis, PITPs are required for both hemostasis and nonhemostatic functions. The discrepancy among the in vivo models suggests that an essential role for PITP-mediated phosphoinositide signaling is not universal and perhaps explains how different biological processes are specialized via compartmentalization of local PITP protein function within platelets. In addition, our previous study14 together with the data shown here, demonstrate that PITP can regulate platelet thrombin generation by supporting the externalization of PS to the platelet surface. This suggests that platelet PITP may play major roles in processes that are highly dependent on thrombin activation, such as tail bleeding and tumor metastasis.
Notably, although murine platelet PITPβ levels are expressed at lower levels than PITPα, PITPβ is just as important as PITPα in phosphoinositide production. Previous studies with N-ethyl maleimide (which blocks PITP opening its hydrophobic pocket) have shown that the rate of lipid exchange by PITPβ is ∼5 times faster than PITPα.28 Experiments with FRET also demonstrated that PITPβ has a higher transfer rate than PITPα; thus, PITPβ may have higher specific activity than the α isoform.29,30 This illustrates that PITPβ could potentially be more efficient than PITPα, requiring less expression. Although PITPs were initially identified for their lipid transfer function, there is growing evidence that they present PtdIns directly to phosphoinositide kinases and therefore have cofactor activity.25,26 For example, Kular et al demonstrated that formylmethionyl-leucylphenylalanine–sensitive PtdIns(3,4,5)P3 production by PtdIns 3–kinase γ in HL60 cells requires PITP to function as a cofactor rather than as a lipid transfer protein.31 Fensome et al found that PITP can restore secretory function in HL60 cells and that PITP promoted PtdIns(4,5)P2 synthesis to restore exocytosis.32 Furthermore, Panaretou et al demonstrated that PITP physically associates with the p150-PtdIns 3–kinase complex in vitro to activate lipid kinase activity.33 Finally, Phillips et al showed that a yeast mutant PITP sec14 that lacked lipid transfer activity could rescue the secretion defects of lethal sec14 null yeast mutations.34
Cockcroft has described PtdIns presentation (cofactor) supplementing the lipid transfer function of PITP.1 Recently, Cockcroft and Atkinson have demonstrated that the PITPβ binds to membrane lipids with higher affinity than PITPα, using dual polarization interferometry.29 It has been proposed that PITP functions as a lipid kinase cofactor by increasing the accessibility of the kinases to the head groups of phospholipids that are normally buried within the lipid bilayer of membranes. Because PITPs prefer highly curved membranes to bind lipids, PITP, once bound to a membrane, may serve as an adapter protein for lipid kinases.17,35,36 Thus, PITP isoforms would interact within a larger protein complex that regulates the lipid kinase machinery.
Although further research is needed to refine these models, our evidence demonstrates that both PITP isoforms regulate phosphoinositide production beyond their traditional transferability role in vivo. Because PITPα and PITPβ differ in their intracellular localization in various tissue cell lines,5,6 this indicates that they may play unique intracellular roles.26 Our data support this model, demonstrating that PITPβ is more evenly distributed in the membrane and cytoskeleton than PITPα, which is almost entirely restricted to the cytosol. We hypothesized that PITPβ functions as a cofactor in local phosphoinositide synthesis at the cell membrane, or perhaps at the cytoskeleton. Furthermore, the data shown here suggest that the loss of either isoform causes spatial redistribution of the other extant isoform, perhaps to maintain basic functional requirements, such as phosphoinositide synthesis. We hypothesized that in platelet phospholipid signaling, PITPα serves the conventional lipid transport function role, whereas both PITPβ and PITPα serve as cofactors in lipid kinases, possibly in different cellular compartments. We further propose that in certain pathways, PITP may operate serially (Figure 7H).
Our study demonstrated that PITP isoforms have overlapping but nonredundant roles in supporting platelet function. Both isoforms contribute to platelet phospholipid signaling and are possibly compartmentalized to impart differential functionality. Despite being significantly less abundant than PITPα, PITPβ contributes to healthy platelet physiology in both hemostatic and nonhemostatic functions.
Acknowledgments
The authors thank Radek Skoda of the University of Basel, Switzerland for providing the PF4-Cre mice used in this study. This work was supported by National Institutes of Health grants PO1 HL120846, PO1 HL146373, 5R01HL148014-02, and PO1 HL40387 (C.S.A.). Phillip T. Hawkins and Len R. Stephens at Signaling ISP, Babraham Institute, Babraham, Cambridge supported mass spectrometry analysis (Biotechnology and Biological Sciences Research Council research grant (BB/PO13384/1).
Authorship
Contribution: K.E.A., B.W., L.Z., C.L.T., A.S., T.J.S., and S.C. performed the experiments; L.Z., C.L.T., A.S., T.J.S., and C.S.A. wrote the manuscript; and L.Z., C.L.T., A.S., T.J.S., S.H.M., S.K., S.C., and C.S.A. designed the experiments, analyzed the data, and reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of S.H.M is Department of Internal Medicine, University of Michigan, Ann Arbor, Michigan.
Correspondence: Charles S. Abrams, Perelman Center for Advanced Medicine, 3400 Spruce St, Room 622, Philadelphia, PA 19104-6160; e-mail: abrams@upenn.edu.
References
Author notes
All data needed to evaluate the conclusions in this paper are present in the paper or the supplemental Materials.
All relevant data are available upon reasonable request from the corresponding author, Charles S. Abrams (abrams@upenn.edu).
The full-text version of this article contains a data supplement.



![Mice lacking platelet PITPs have prolonged tail bleeding time, but no defects in laser-induced in vivo thrombosis. The laser-induced injury model demonstrates normal in vivo thrombosis and platelet secretion in the Pitpαfl/fl/βfl/flPf4-Cre+ mice. (A) Representative images show platelet accumulation (CD41, red) and P-selectin exposure (green [overlay of red/green is yellow]) 3 minutes after laser-induced injury to the cremaster arterioles. Images are binary representations of 2D confocal fluorescence images overlaid on the bright-field. White arrows indicate the direction of flow; scale bar, 10 μm. (B) Graphs of the CD41+ area over time (left, mean ± standard error of the mean), median CD41+ area over time (middle), and CD41 peak area (right, lines represent median ± interquartile range). (C) Graphs of the P-selectin–positive area over time (left, mean ± standard error of the mean), the median P-selectin–positive area over time (middle), and P-selectin peak area (right, lines are median ± interquartile range). n = 20 thrombi in 4 wild-type mice and n = 29 thrombi in 4 Pitpαfl/fl/βfl/flPf4-Cre+ mice. Statistical analysis were performed using a two-tailed Mann-Whitney test. (D,E) Tail bleeding times in mice lacking PITPβ (D) or both PITP isoforms compared with their littermate controls (E). Tail bleeding was normal in mice with platelets lacking PITPβ (NS, Mann-Whitney test; n = 52 for Pitpβfl/flPf4-Cre- mice and n = 58 for Pitpβfl/flPf4-Cre+ mice). When both PITP isoforms were deleted in platelets, there was a mild increase in bleeding time (P = .0013 using two-tailed Mann-Whitney test; n = 57 for Pitpαfl/fl/βfl/flPf4-Cre- mice and n = 54 for Pitpαfl/fl/βfl/flPf4-Cre+ mice).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/16/10.1182_bloodadvances.2022008735/2/m_blooda_adv-2022-008735-gr2.jpeg?Expires=1765937110&Signature=KKNBFQDBYU4ds99KYufwqMhepp4bwhAQ-jaoDGNq9kQ8bzy1N69ZlBmY2Ek3hR827mkt~2hpPVqTLuilyhVv25UHdUH4rkXbqRhc4p-qzBzTCe2zcYZmd9qjOUQp6r7aouHqXFRIzq7dEz5oWxj1Xeb0BpPN89pp1HKaIq5TV0O-qTAcDcARPhVivlcPj4exGbqrIll1qT43S5HPLBCPkhagJYqrnsG2bUYqCu4U0RoVf3XUz4UW65yEvfHWC8zr3eOJvH15ZxNRzs~e0WBqb17Km5EWD-aHD~jR1uCx3yzdspB0BHtnxABkakpqebWvIC16xZgSIFC~6po~iQzAtg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![PITPβ does not have transfer activity but has cofactor activity. (A) In vitro [3H]-labeled PtdIns transfer activity from microsomes (permeabilized HL60 cells) to liposomes (PC:PI :: 98:2) is mediated by platelet PITPα, but not PITPβ. (B.C) Lipid kinase assays were performed to determine the effects of platelet PITPα (left) and PITPβ (right) on PtdInsP synthesis in vitro, before and after thrombin stimulation (3 minutes: time of thrombin stimulation [1 U/mL]). This assay, which does not require transfer activity, demonstrated that both PITP isoforms are required for phospholipid kinases to generate phosphoinositides. Phosphoinositide production in PITP-null platelets was restored by the addition of rPITPα and rPITPβ. ∗∗P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/16/10.1182_bloodadvances.2022008735/2/m_blooda_adv-2022-008735-gr6.jpeg?Expires=1765937110&Signature=eGL5IMhFM8XKMYRBW3Zlezn2fGE7gRiRNRC9GyvhgWR-RCOZXd01BHUlEQy2m-~tUF1z0QmEWGkZiPvbKmcXphbFoXeMlPJ6~jx0AY7MwQMGqLQYEiZOsdCa4f4iyoW50n-ft4I~YmdBRRG8b8xlaDlp-dRa8mgy-HwbC~UDu5anL55eVVh40100dKfLH86tGXav39RrarQ6oi9E6s3tZykauYby0fo4PWGRJLCcA9tkImnW26K55l5FGvozFrHzuJ6N2df9JvOhhSqkkGqST-bd94d0CzHibCtx-3qgbWWHKfXHGnxE9tjEPVL8r9Y8RAwwHb5zwNQlEzckFgnXUQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Mice lacking platelet PITPs have prolonged tail bleeding time, but no defects in laser-induced in vivo thrombosis. The laser-induced injury model demonstrates normal in vivo thrombosis and platelet secretion in the Pitpαfl/fl/βfl/flPf4-Cre+ mice. (A) Representative images show platelet accumulation (CD41, red) and P-selectin exposure (green [overlay of red/green is yellow]) 3 minutes after laser-induced injury to the cremaster arterioles. Images are binary representations of 2D confocal fluorescence images overlaid on the bright-field. White arrows indicate the direction of flow; scale bar, 10 μm. (B) Graphs of the CD41+ area over time (left, mean ± standard error of the mean), median CD41+ area over time (middle), and CD41 peak area (right, lines represent median ± interquartile range). (C) Graphs of the P-selectin–positive area over time (left, mean ± standard error of the mean), the median P-selectin–positive area over time (middle), and P-selectin peak area (right, lines are median ± interquartile range). n = 20 thrombi in 4 wild-type mice and n = 29 thrombi in 4 Pitpαfl/fl/βfl/flPf4-Cre+ mice. Statistical analysis were performed using a two-tailed Mann-Whitney test. (D,E) Tail bleeding times in mice lacking PITPβ (D) or both PITP isoforms compared with their littermate controls (E). Tail bleeding was normal in mice with platelets lacking PITPβ (NS, Mann-Whitney test; n = 52 for Pitpβfl/flPf4-Cre- mice and n = 58 for Pitpβfl/flPf4-Cre+ mice). When both PITP isoforms were deleted in platelets, there was a mild increase in bleeding time (P = .0013 using two-tailed Mann-Whitney test; n = 57 for Pitpαfl/fl/βfl/flPf4-Cre- mice and n = 54 for Pitpαfl/fl/βfl/flPf4-Cre+ mice).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/16/10.1182_bloodadvances.2022008735/2/m_blooda_adv-2022-008735-gr2.jpeg?Expires=1766543663&Signature=yzdrmZ-EUZqbN8m-GIoc38KEqfR3Hk8DrCFgv05AcXr2jh85mAKUS4mh~f3meGcSnozkBHzc9304KV4Grjlr1VLzJl2E3wBHvQk7hp-lp~0ObwVOcOdBPWkryGMbwbSyQ6sI5ci-HADFWLDBlhgzxCMUNfavQg~9BeMpFiXFJ8HEYi8p8-LkjDZW~4FGR9nLY5wK7AS3tFVfUmfDgYsM9iXWu9Qvbul4IJExnPsTEOLCRdLVg-uDW23i3E4bvDMcC1TstErLgPyuJ6pQN2~wd2Dna6E8uBu4bWOz9WDcdhf5cJ5ok2bV27JuhV7WnaR6cfzD4Uy63bNk2Ebu3qioQQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![PITPβ does not have transfer activity but has cofactor activity. (A) In vitro [3H]-labeled PtdIns transfer activity from microsomes (permeabilized HL60 cells) to liposomes (PC:PI :: 98:2) is mediated by platelet PITPα, but not PITPβ. (B.C) Lipid kinase assays were performed to determine the effects of platelet PITPα (left) and PITPβ (right) on PtdInsP synthesis in vitro, before and after thrombin stimulation (3 minutes: time of thrombin stimulation [1 U/mL]). This assay, which does not require transfer activity, demonstrated that both PITP isoforms are required for phospholipid kinases to generate phosphoinositides. Phosphoinositide production in PITP-null platelets was restored by the addition of rPITPα and rPITPβ. ∗∗P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/16/10.1182_bloodadvances.2022008735/2/m_blooda_adv-2022-008735-gr6.jpeg?Expires=1766543663&Signature=xVxypKYzaPlAYupUx2no7ELpVFzBsBrBMx4yPzdgOtVQdoHclQgadpin1sKHlygUThqPkHDcCQLo817uSdAVpOGvUpsrWsPIpWsCLZL8xKmjXfbxs0FfhaD2Cqra6IyzOT5lQSDsOZxXpf1BowUMEOTvOlvrbRFQEPr~XRgKojHGDei8JZ99jBGAeUTpmHaE4HnPf8j6ldYykCJUDPnVIPMlMXAkE9iCTqeimjwjCCHGYClut1R0qXrgnefOt1FRI~h4uoCo41eoIs-Tel3V6oq3EEwFo2RAmaofra0jmsHhUCkCTWmQNNnc3GOm03wZP8w2ouj0b9LSlVROOfHReA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
