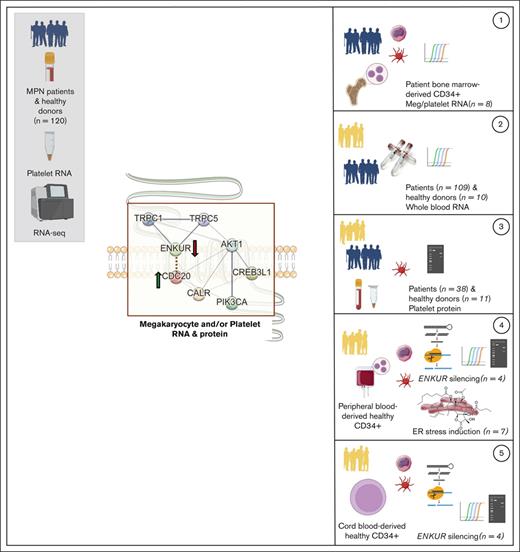
Key Points
Enkurin, a calcium channel adapter protein, is identified as a novel marker of pathogenesis in MPN.
MPN megakaryocyte and platelet expression of ENKUR at RNA and protein levels suggest ER stress/calcium dysregulation in MPN pathogenesis.
Abstract
Impaired protein homeostasis, though well established in age-related disorders, has been recently linked with the pathogenesis of myeloproliferative neoplasms (MPNs). However, little is known about MPN-specific modulators of proteostasis, thus impeding our ability for increased mechanistic understanding and discovery of additional therapeutic targets. Loss of proteostasis, in itself, is traced to dysregulated mechanisms in protein folding and intracellular calcium signaling at the endoplasmic reticulum (ER). Here, using ex vivo and in vitro systems (including CD34+ cultures from patient bone marrow and healthy cord/peripheral blood specimens), we extend our prior data from platelet RNA sequencing in patients with MPN and discover select proteostasis-associated markers at RNA and/or protein levels in each of platelet, parent megakaryocyte, and whole blood specimens. Importantly, we identify a novel role in MPNs for enkurin (ENKUR), a calcium mediator protein originally implicated only in spermatogenesis. Our data reveal consistent ENKUR downregulation at both RNA and protein levels across specimens from patients with MPN and experimental models (including upon treatment with thapsigargin, an agent that causes protein misfolding in the ER by selective loss of calcium), with a concomitant upregulation of a cell cycle marker, CDC20. Silencing of ENKUR using short hairpin RNA in CD34+–derived megakaryocytes further confirms this association with CDC20 at both RNA and protein levels and indicates a likely role for the PI3K/Akt pathway. Together, our work sheds light on enkurin as a novel marker of MPN pathogenesis and indicates further mechanistic investigation into a role for dysregulated calcium homeostasis and ER and protein folding stress in MPN transformation.
Introduction
Myeloproliferative neoplasms (MPNs) are a group of malignant disorders of the bone marrow in which a dysregulated balance between proliferation and differentiation gives rise to abnormal numbers of blood cells in circulation. Classical MPNs1,2 are defined by a combination of clinical, laboratory, morphological, and molecular genetic features and constitute 3 phenotypic subtypes: essential thrombocythemia (ET), polycythemia vera (PV), and myelofibrosis (MF; primary or secondary, if transformed from prior ET/PV). Somatic mutations in 1 of 3 driver genes (JAK2, CALR, and MPL) constitute their shared molecular genetic pathogenesis, causing constitutive JAK/STAT signaling in hematopoietic stem cells.3-7 The mutated CALR also induces oncogenic effects by binding and constitutively activating the thrombopoietin receptor and the downstream signaling cascade,7,8 causing cellular transformation and abnormal megakaryopoiesis. To date, therapeutic strategies have largely focused on JAK2 inhibition (eg, ruxolitinib [RUX]9) and are effective in alleviating MPN symptoms, but only partially, given their primary role in suppression of inflammation and reduction of circulating proinflammatory cytokines.10,11 Additional therapeutic strategies that target MPN pathogenetic mechanisms12-17 would be critical in resolving patient disease burden.
A recent study18,19 assessing mechanisms of MPN pathogenesis identified dysregulation of proteostasis and protein quality control at the endoplasmic reticulum (ER) as crucial transformative events and therefore a strong therapeutic target. However, this study was primarily in mice and remains to be confirmed in independent additional MPN models and patient-derived specimens. Our recent investigation, profiling the blood platelet transcriptome in all 3 subtypes of patients with MPN20 identified high expression of genes and pathways associated with impaired proteostasis, ER stress, and unfolded protein response (UPR), which is well recognized in other age-related disorders.21-25 Here, we extend our prior platelet transcriptomic data of patients with MPN, with a focus on genes associated with proteostasis, cell proliferation, ER stress, or calcium signaling at high statistical significance (false discovery rate < 0.01) and representing 4 types of trends in platelet RNA expression across MPN subtypes: progressively downregulated, progressively upregulated, consistently upregulated, and uniquely upregulated in MF alone. Genes selected include (1) ENKUR (enkurin),26 a little-studied transient receptor potential cation channel27,28 adapter protein with known function primarily in the context of sperm motility alone,26,29 (2) CALR, a well-known ER chaperone and calcium-binding protein,30-33 also relevant in the context of megakaryocyte (MK) to platelet differentiation,34-37 (3) cyclic adenosine monophosphate–responsive element-binding protein CREB3L1, an ER-Golgi stress transducer and transcription protein involved in the UPR response,38-41 and (4) CDC20, involved in cell division and proliferation42-44 and known to be upregulated in other cancers.44 Using patient bone marrow–, healthy cord–, and peripheral blood–derived CD34+ cells differentiated into MK and platelet cultures ex vivo, we consistently identify ENKUR as a novel peripheral and marrow biomarker in MPNs at both RNA and protein levels. Our data also demonstrate a negative correlation of ENKUR with CDC20 expression in MK/platelet cultures and in independent banked MPN whole blood RNA specimens. Beyond the utility of enkurin as a potential marker of chronic vs advanced MPN subtypes, more research is warranted to elucidate the mechanisms of its downregulation in MPNs and the consequent targeting of cell proliferation pathways.
Materials and Methods
Please see Table 1 for a tabulated overview of all RNA and protein measurements in this study.
We collected bone marrow specimens from patients with MPN from the Stanford Cancer Center and whole blood from healthy donors at the Stanford Blood Center. Cell culture supplies, including plates, fetal bovine serum, penicillin, streptomycin, phosphate-buffered saline (PBS), and culture medium, were procured from GIBCO BRL (Frederick, MD). Antibodies for CALR, CDC20, p-PI3K, p-Akt, total PI3K, total Akt, β-Actin, and anti–rabbit goat immunoglobulin G were procured from Cell Signaling Technologies (Danvers, MA), for enkurin from Sigma Aldrich (St Louis, MO), and for CREB3L1 from Thermo Fisher Scientific (Rockford, IL). Antibodies for select cell markers conjugated with fluorophores were procured from BioLegend (San Diego, CA) and constituted CD41 (Alexa647, red-blue), CD42b (PE, yellow-blue), CD61 (PE), and CD45 (PE).
Short hairpin RNA (shRNA) for ENKUR silencing was synthesized at Protein and Nucleic acid facility at Stanford University (Stanford, CA). The human HEK293 FT cells and THP1 human macrophage cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured following the supplier instructions. The CD34 and CD45 microbeads, LS and MS columns, and magnetic separators were purchased from Miltenyi Biotech (Cambridge, MA). Cytokines interleukin-6, thrombopoietin (TPO), and Flt3 ligand were from PeproTech Inc (Cranbury, NJ); stem cell factor (SCF) and SFEM II media were from Stem Cell Technologies (Kent, WA). Lenti-X GoStix Plus was purchased from TaKaRa Bio Inc (San Jose, CA).
Peripheral blood platelet isolation
Peripheral blood was collected in acid citrate-dextrose (3.2%) sterile yellow-top tubes (Becton, Dickinson and Co) and was processed within 4 hours of collection for all samples. Platelets were isolated by established45-48 purification protocols. Briefly, the acid citrate-dextrose tube whole blood was first centrifuged at 200 g for 20 minutes at room temperature. The platelet-rich plasma (PRP) was removed, and prostaglandin E1 was added to the PRP to prevent exogenous platelet activation. The PRP was then centrifuged at 1000 g for 20 minutes at room temperature. The platelet pellet was resuspended in warmed (37°C) PIPES saline glucose. Leukocytes were depleted using CD45+ magnetic beads (Miltenyi Biotec). Isolated platelets were further resuspended in Trizol or LDS buffer for RNA (polymerase chain reaction [PCR]) and protein (western blot) analyses.
RNA extraction and quantification from whole blood (PAXgene tubes)
Whole blood from patients with MPN and healthy donors was collected into PAXgene tubes (BD Biosciences) containing red blood cell lysis buffer. RNA was isolated using PAXgene Blood RNA kit, following the manufacturer’s instructions (762164, PreAnalytiX, Hombrechtikon, Switzerland).
CD34+ cell isolation from MPN patient bone marrow and healthy peripheral and cord blood cultures
CD34+ cells were isolated49,50 using microbead positive selection (Miltenyi Biotec) from source specimens of peripheral blood or cord blood of healthy donors or bone marrow of patients with MPN (collected as part of their clinical care with research consent). Briefly, the RBCs were lysed using RBC lysis buffer, and the cells were pelleted. Cells were suspended in MACS running buffer, incubated with the CD34+ microbeads for 1 hour on ice, and then passed through MACS LS columns pre-equilibrated with MACS running buffer. CD34+ cells thus collected were washed using MACS running buffer and resuspended in the SFEM II media (4 × 105 to 5 × 105 cells per mL) containing TPO (20 ng/mL), SCF (25 ng/mL), and gentamicin (1:1000) and plated/transferred to 12-well plates at 37°C in 5% CO2. On day 3, cells were resuspended in fresh media (SFEM II) containing TPO (20 ng/mL), SCF (25 ng/mL), and gentamicin (1:1000), and on days 6, 9, and 12, in TPO (40 ng/mL) and gentamicin (1:1000). The cells were harvested on day 15. Cells were collected and centrifuged at 300 g for 10 minutes to pellet the larger MK fraction and at 3000 g for 30 minutes for the smaller platelet fraction. With the healthy peripheral- and cord blood–derived CD34+ cells, the isolated cells were expanded using hematopoietic stem cell expansion media (Cell Genix) containing 100 ng/mL of interleukin-6, Flt3 ligand, SCF, and TPO for 5 days before starting the culture. The formation of MK cells and platelets was confirmed via surface marker analysis, using CD41, CD42b, and CD61.
All samples from patients with MPN and healthy donors were obtained after written informed patient consent and were fully anonymized. Study approval was provided by the Stanford University institutional review board. All relevant ethical regulations were followed.
Cell-surface marker analysis using flow cytometry
After isolation of the MK and platelet fractions from the 15-day (+ TPO) CD34+ culture, cells were collected and washed with PBS. For the CD41, CD42b, and CD61 stains, cells were resuspended in MACS running buffer and stained with Alexa 647 anti-human CD41, PE anti-human CD42b, and PE anti-human CD61 and incubated at room temperature, protected from light for 45 minutes. Cells were washed with PBS and resuspended at a final concentration of 2 × 107 cells per mL in PBS before running the sample on the flow cytometer. All experiments were performed using GUAVA flow cytometer, and all flow cytometry analysis was performed in the FlowJo Software. Data from flow cytometry experiments was acquired by gating for events that were in focus.
Total RNA isolation and PCR with CD34+–derived MK and platelet fractions
Total RNA was isolated from the MK and platelet fractions using a mirVana RNA extraction kit (Life Technologies, Grand Island, NY) in accordance with the manufacturer’s instructions. Briefly, MKs/platelets were homogenized in 300 μL lysis buffer, then incubated with a 30 μL homogenate additive for 10 minutes. RNA was extracted with acid-phenol and column-purified, then washed 3 times with washing buffer and eluted in 40 μL of sterile elution buffer. The total RNA was quantified using a Nanodrop spectrophotometer, and then 100 ng of total RNAs were reverse-transcribed using a Reverse-Transcription Kit (Life Technologies) and reverse transcriptase (RT) primers to synthesize complementary DNA. Then, quantitative RT-PCR (qRT-PCR) was performed using the TaqMan-PCR primers and probe mix combined with the complementary DNA derivates. qRT-PCR was performed through a 2-minute incubation at 50°C, followed by DNA polymerase activation at 95°C for 10 minutes, plus 60 cycles at 95°C for 15 seconds and 60°C for 60 seconds in the BioRad CFX96 thermocycler system (BioRad). The qRT-PCR reaction procedure was executed in a 20 μL final reaction volume. The expression of target genes was analyzed using the 2−ΔΔCt method.
Thapsigargin-induced ER stress
Isolated human CD34+ cells from the peripheral blood of healthy volunteers were cultured for 15 days with the supplementation of both SCF and TPO for the first 6 days and then with TPO alone until day 15. Fresh media containing either SCF and/or TPO was added every 3 days. To induce ER stress in these cells, the known ER stressor drug thapsigargin (125 nM, as optimum dosage was identified via preliminary experiments) was added to the culture on day 7. The culture was maintained until day 15, with media change every 3 days. The cells were harvested and analyzed for gene expression and immunoblotting for the target proteins.
Western blotting for protein expression in MKs and platelets
Isolated megakaryocytes (MKs) were aliquoted for flow cytometry, RNA expression, and western blotting. For western blotting, an aliquot of the MK cells was pelleted and lysed using cell lysis buffer containing protease and phosphatase inhibitor cocktails and EDTA. The samples were then resolved on a reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (10% acrylamide). Blots were stained using appropriate primary (anti-human rabbit antibody; 1:1000 volume-to-volume ratio) and secondary antibodies (anti-rabbit immunoglobulin G horseradish peroxidase conjugate; 1:2000 volume-to-volume ratio).
Protein expression was determined based on the detection of a band. The intensity of the protein bands observed was semiquantified using IVIS or ImageJ software, with normalization of each protein against β-actin. Blots were visualized using enhanced chemiluminescent (ECL) reagent via the Amersham Imager 680 or by IVIS imaging systems.
ENKUR gene silencing
Construction of the vector
PLKO1 vector was used to construct the ENKUR-silenced cell lines in HEK293T cells (known for their ease of transfection and fast growth rate). The PLKO1 vector was cut using EcoR1 and Age1 to ligate the shRNA using T4 ligase.
Two independent shRNA sequences were used to knock down ENKUR and contrasted with a scramble sequence; all 3 sequences are shown below:
shRNA1: PCCGGTCCGGCCAACCTCGATACTCTTATTTCTCGAGAAATAAGAGTATCG
AGGTTGGTTTTTTTTGG
shRNA2:
PCCGGTCCGGCATGGGAGTGGCTAAAAAGCCCTCGAGGGCTTTTTAGCC
ACTCCCATGTTTTTTTTGG
Scrambled: PCCGGTCCGGGTGCGTTGCTAGTACCAACTCTCGAGAGTTGGTACTA
GCAACGCACTTTTTTTTGG
Validation of the construct
The silencing effect of the shRNA construct was assessed using HEK293 FT cells via the transfection of the shRNA-containing plasmids (pLKO1-shRNA), using the lipofectamine method. Briefly, 100K HEK293 FT cells were seeded in a 12-well plate 24 hours before the transfection. Transfection was performed by adding a master mix containing lipofectamine and the shRNA to reduced serum media for 4 hours before adding the complete media. The cells were harvested 48 hours after transfection, and the silencing effect was assessed via western immunoblotting against enkurin.
Cell culture for ENKUR silencing
HEK293 FT cells were cultured in Dulbecco’s modified Eagle medium/high glucose with 10% fetal bovine serum, 0.1% streptomycin, and 100 U/mL penicillin at 37°C, 5% CO2, and 95% air environment, whereas THP-1 cells were cultured in RPMI media containing 10% fetal bovine serum, 0.1% streptomycin, and 100 U/mL penicillin at 37°C, and 5% CO2, environment. The cells were tested for any mycoplasma contamination using a MycoAlert kit (Lonza, Allendale, NJ) and maintained at optimum cultural conditions.
Production of lentiviral particles
After confirmation of the silencing effect of the shRNA construct, it was packaged along with the viral plasmids to produce the lentivirus. Briefly, the lentiviral vectors were transfected into HEK293 FT cells using the calcium phosphate method. The cells were supplemented with chloroquine and N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid. After 48 hours of incubation, the media was carefully aspirated, and the viral particles were concentrated via ultracentrifugation. The infectious units (IFU/ml) value was determined using Lenti-X GoStix, following the manufacturer’s instructions (Cat#631280).
Validation of the lentivirus silencing in HEK cells
As described earlier, 100K HEK293 FT cells were plated in a 12-well plate 24 hours before lentiviral infection (transduction). The media was aspirated, and a lentivirus master mix containing polybrene in reduced serum media was added to the cells. After 4 hours of incubation, the cells were supplemented with complete media and incubated for 48 hours before harvesting. The cells were lysed and immunoblotted against enkurin to assess the gene silencing effect.
ENKUR gene silencing in CD34+–derived MKs
The shRNA-expressing lentiviral particles were infected into the CD34+ cells in culture on day 9 (MKs derived from CD34+ cells) at a ratio of 1:20 (cell–to–viral particle ratio). The culture was maintained for another 6 days, with TPO supplementation every 3 days. Cells were harvested on day 15 and centrifuged at 2 speeds: low (300 g) and high (3000 g) to separate the MK and platelet fractions, respectively. Each fraction was divided into 3 aliquots for assessing each of the cell-surface markers (50 × 103-100 × 103 cells), RNA (∼50 × 103 cells), and protein (100 × 103-200 × 103 cells) expression.
Statistical analyses
Continuous variables from all experiments were assessed for normality. Data that were normally distributed were expressed as the mean ± the standard error of the mean. For analyses involving 2 groups, a parametric two-tailed Student t test was used. When 3 or more groups were analyzed, an analysis of variance with a Tukey post hoc test was performed. When data were not normally distributed, a Mann-Whitney was used when 2 groups were analyzed, whereas a Kruskal-Wallis with a Dunn multiple comparison post hoc test was used for analyses of 3 or more groups. When appropriate, a two-way analysis of variance with a post hoc test was used as described. Summary statistics were used to describe the study cohort, and clinical variables were expressed as the mean ± error of the mean or as a number and percentage (%). Statistical analyses were performed by using Prism (version 9; GraphPad, San Diego, CA), and a P value < .05 was considered statistically significant.
Results
Validation of RNA and protein expression of 4 MPN candidate markers
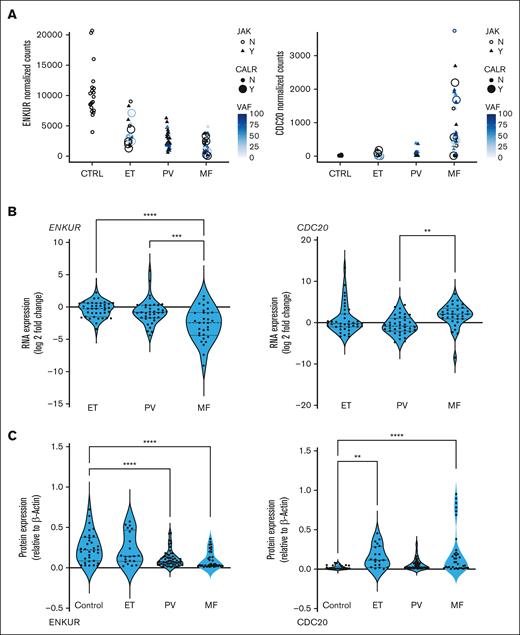
Four candidate markers from our prior platelet RNA sequencing (RNA-seq) analyses20 were identified based on specific trends in progressive expression across the 3 MPN subtypes vis-à-vis healthy donors (false discovery rate < 0.01; n = 120), independent of patient driver mutational status (and allelic burden). ENKUR and CALR, in particular, were identified based on opposing trends in progressive expression (Figure 1A; supplemental Figure 1A), CDC20 based on its specific high expression in patients with MF alone in contrast with healthy donors and patients with ET or PV (Figure 1A, top right), and CREB3L1 based on its substantially increased (>50-fold) expression in all MPNs vs healthy (supplemental Figure 1A, top right). Given that the platelet RNA profile is a composite of influences from the peripheral circulation as well as the genetic (and MPN mutational) profile of MKs in the bone marrow, we hypothesized a priori that our MPN candidate markers are likely to be variably validated in one vs the other.
RNA expression in platelets and whole blood of patients with MPN and protein expression in platelets. (A) MPN platelet RNA-seq normalized expression data: patients with MPN (total, n = 99; ET, n = 24; PV, n = 33; and MF, n = 42) vs healthy donors (n = 21) annotated further by (i) shape for JAK2 mutation status (triangle = yes and open circle = no), (ii) filled circle size for CALR mutation status, and (iii) color for variant allele fraction (VAF) for each patient. (B) Whole blood RNA: patients with MPN (total, n = 109; ET, n = 39; PV, n = 38; and MF, n = 32) vs healthy donors (n = 10). (C) Platelet protein: peripheral blood platelets were isolated from patients with MPN (total, n = 38; MF, n = 12; PV, n = 19; and ET, n = 7) and healthy donors (n = 11). Bonferroni multiple comparisons adjusted P values, ∗∗∗∗P < .0001; ∗∗∗P < .001; and ∗∗P < .01 when compared with controls (healthy donors). All RNA expression is normalized to glyceraldehyde-3-phosphate dehydrogenase and expressed as a log2 fold change.
RNA expression in platelets and whole blood of patients with MPN and protein expression in platelets. (A) MPN platelet RNA-seq normalized expression data: patients with MPN (total, n = 99; ET, n = 24; PV, n = 33; and MF, n = 42) vs healthy donors (n = 21) annotated further by (i) shape for JAK2 mutation status (triangle = yes and open circle = no), (ii) filled circle size for CALR mutation status, and (iii) color for variant allele fraction (VAF) for each patient. (B) Whole blood RNA: patients with MPN (total, n = 109; ET, n = 39; PV, n = 38; and MF, n = 32) vs healthy donors (n = 10). (C) Platelet protein: peripheral blood platelets were isolated from patients with MPN (total, n = 38; MF, n = 12; PV, n = 19; and ET, n = 7) and healthy donors (n = 11). Bonferroni multiple comparisons adjusted P values, ∗∗∗∗P < .0001; ∗∗∗P < .001; and ∗∗P < .01 when compared with controls (healthy donors). All RNA expression is normalized to glyceraldehyde-3-phosphate dehydrogenase and expressed as a log2 fold change.
Firstly, in whole blood RNA of patients with MPN vs healthy donors, expression of ENKUR and CDC20 was confirmed via qPCR (Figure 1B; n = 109; P < .005) as nearly sixfold downregulated and threefold upregulated, respectively, in the advanced MPN subtype, MF. Expression of CREB3L1 was also confirmed as more than 15-fold upregulated across ET, PV, and MF (supplemental Figure 1B, right panel), whereas expression of CALR (supplemental Figure 1B, left panel) was variable and not uniformly consistent with the platelet transcriptome (likely because of lower resolution for detection of transcripts in the whole blood). Next, in assessing platelet protein levels, expression of ENKUR was significantly different in patients with PV and MF vs that in healthy donors, and expression of CDC20 differed in patients with ET and MF (Figure 1C), whereas CALR and CREB3L1 were not significantly different between patient and healthy donor specimens (supplemental Figure 1C).
MK fractions from CD34+ cultures of bone marrow from patients with MPN
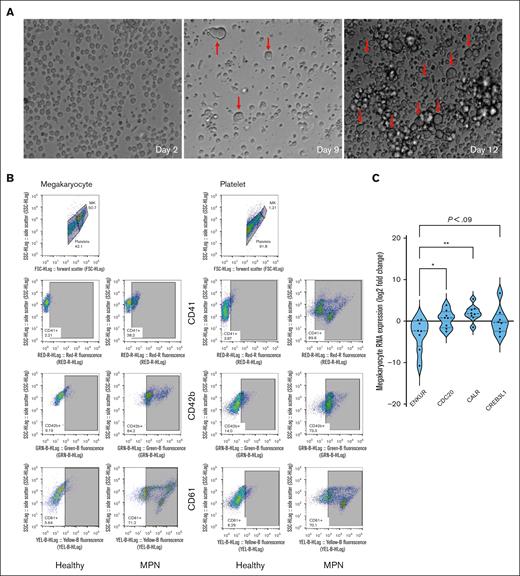
To further delineate RNA expression of the candidate markers as specific to platelets in circulation alone or also in marrow-derived MKs, we established an ex vivo culture of CD34+ stem cells from bone marrow of patients with MPN (n = 8; MF, 4) and generated MK and platelet fractions (Figure 2A-B; see “Materials and Methods” and supplemental Figure 2A-B for flow cytometric and histological confirmation of cultured MK and platelet fractions). Given the lower cell quantity of the nascent platelet fraction than the megakaryocytic fraction, we focused entirely on the MKs harvested. Downregulation of ENKUR was recapitulated in these patient-derived megakaryocytic RNA (mean, approximately twofold reduction in MF alone and fivefold all MPN), with associated upregulation of CDC20, CREB3L1 (mean, approximately onefold), and CALR (approximately fourfold) in all MPNs (n = 8; Figure 2C).
Culturing of CD34+ cells from the bone marrow of patients with MPN and RNA expression. (A) Microscopic images of the CD34+ cells isolated from the bone marrow of patients with MPN over the 15-day culture period at original magnification ×20. Notice the formation of MKs indicated with a red arrow. (B) Flow cytometric analysis of cell-surface markers to confirm the production of MK and platelet fractions after 15 days of culture of CD34+ cells from healthy cord or peripheral blood or MPN patient bone marrow sources. (C) Megakaryocytic RNA: cell culture from fresh patient bone marrow–derived CD34+ cells then differentiated over 15 days into MKs (n = 8). All RNA expression is normalized to glyceraldehyde-3-phosphate dehydrogenase and expressed as a log2 fold change. Bonferroni adjusted P values, ∗∗P < .01; ∗P < .05.
Culturing of CD34+ cells from the bone marrow of patients with MPN and RNA expression. (A) Microscopic images of the CD34+ cells isolated from the bone marrow of patients with MPN over the 15-day culture period at original magnification ×20. Notice the formation of MKs indicated with a red arrow. (B) Flow cytometric analysis of cell-surface markers to confirm the production of MK and platelet fractions after 15 days of culture of CD34+ cells from healthy cord or peripheral blood or MPN patient bone marrow sources. (C) Megakaryocytic RNA: cell culture from fresh patient bone marrow–derived CD34+ cells then differentiated over 15 days into MKs (n = 8). All RNA expression is normalized to glyceraldehyde-3-phosphate dehydrogenase and expressed as a log2 fold change. Bonferroni adjusted P values, ∗∗P < .01; ∗P < .05.
ENKUR gene silencing
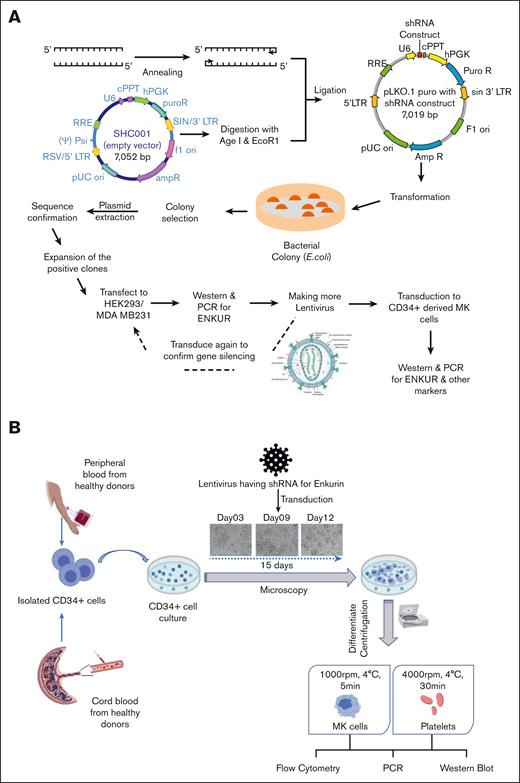
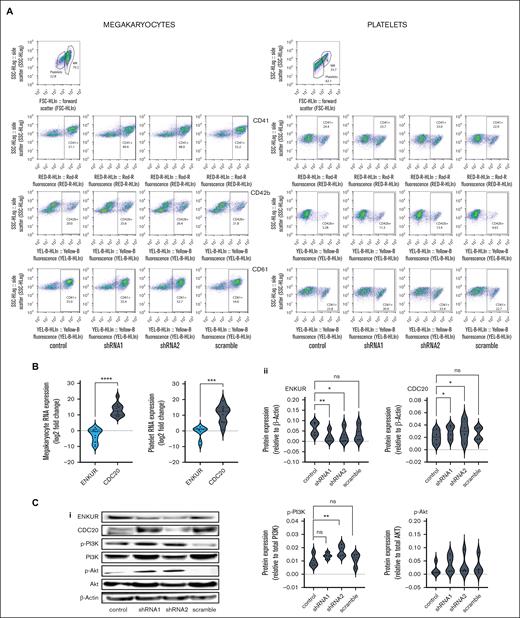
To better evaluate the negative correlation between ENKUR and CDC20/CALR expression, we sought lentiviral knockdown in primary CD34+ cells (derived from healthy peripheral blood and cord blood specimens), using 2 distinct ENKUR shRNA constructs (see Methods). Figure 3A-B describe our experimental framework. Firstly, the shRNAs were expressed in pLKO.1 plasmid that was amplified and isolated from Escherichia coli, and the lentiviral plasmids were then transfected into HEK293 FT cells to generate the lentivirus. ENKUR shRNA silencing and downregulation of enkurin protein were confirmed at a concentration of 1:20 HEK cell–to–viral particle ratio. Primary CD34+ cells from healthy peripheral (n = 4) and cord blood (n = 4) specimens were cultured in 4 conditions: (1) control with no ENKUR silencing, (2) and (3) lentiviral knockdown with the 2 shRNA constructs, and (4) scramble control shRNA. When relevant, shRNA infection was introduced on day 7 of culture and continued for another 7 days (totaling as a 15-day culture; Figure 3B). MK and platelet fractions thus derived were confirmed via flow cytometric analysis (Figure 4A). Downregulated RNA expression of ENKUR (Figure 4B; log2FC = −2.82 ± 1.27) was significantly associated with concomitant high expression of CDC20 (Figure 4B; log2FC = 13.14 ± 1.2) in both of the MK and platelet fractions. Cell quantity in the platelet fraction was just sufficient for RNA qPCR measurements alone so protein levels could be assessed only in the MKs. Inverse association of expression between ENKUR and CDC20 was validated even at megakaryocytic protein levels (Figure 4C; adj P < .05 for each control vs shRNA). However, ENKUR silencing alone (without the associated patient factors in our in vitro experiments, eg, mutational status) was insufficient to generate a statistically significant differential in CALR or CREB3L1 expression at both RNA and protein levels (supplemental Figure 3A-B).
Experimental design and workflow of lentiviral transduction–induced silencing of the ENKUR gene in CD34+ stem cells. (A) Workflow of the shRNA construction and generation of the lentivirus. (B) Experimental plan for the silencing of the ENKUR gene in CD34+ stem cells. The CD34+ cells were isolated from both peripheral and cord blood.
Experimental design and workflow of lentiviral transduction–induced silencing of the ENKUR gene in CD34+ stem cells. (A) Workflow of the shRNA construction and generation of the lentivirus. (B) Experimental plan for the silencing of the ENKUR gene in CD34+ stem cells. The CD34+ cells were isolated from both peripheral and cord blood.
Lentiviral transduction–induced silencing of ENKUR gene in CD34+ stem cells. (A) Flow cytometric analysis of cell-surface markers to confirm the megakaryocytic and platelet fractions after 15 days of culture. (B) RNA expression levels in the CD34+ cell–derived MKs and platelets. (C) (i) Western blot of CD34+-derived MKs (n = 4 cord blood and n = 4 peripheral blood); (ii) densitometry for the blots. shRNA1 and shRNA2 indicate the 2 shRNA sequences used to silence enkurin (”Materials and Methods”), with a scramble sequence and no silencing as negative and positive controls, respectively. Densitometry was performed using the IVIS imaging software (∗∗∗∗P < .0001; ∗∗∗P < .001; ∗∗P < .01; ∗P < .05, when compared with nonsilenced controls).
Lentiviral transduction–induced silencing of ENKUR gene in CD34+ stem cells. (A) Flow cytometric analysis of cell-surface markers to confirm the megakaryocytic and platelet fractions after 15 days of culture. (B) RNA expression levels in the CD34+ cell–derived MKs and platelets. (C) (i) Western blot of CD34+-derived MKs (n = 4 cord blood and n = 4 peripheral blood); (ii) densitometry for the blots. shRNA1 and shRNA2 indicate the 2 shRNA sequences used to silence enkurin (”Materials and Methods”), with a scramble sequence and no silencing as negative and positive controls, respectively. Densitometry was performed using the IVIS imaging software (∗∗∗∗P < .0001; ∗∗∗P < .001; ∗∗P < .01; ∗P < .05, when compared with nonsilenced controls).
One classical downstream pathway linked previously to the ER stress response18,19 in MPNs51 and other cancers52-54 is the PI3K/Akt signaling cascade. Here, with shENKUR, we briefly evaluated a possible association with the PI3K/Akt pathway and found that silencing of enkurin correlated with increased expression of phosphorylated PI3K in megakaryocytes (Figure 4C).
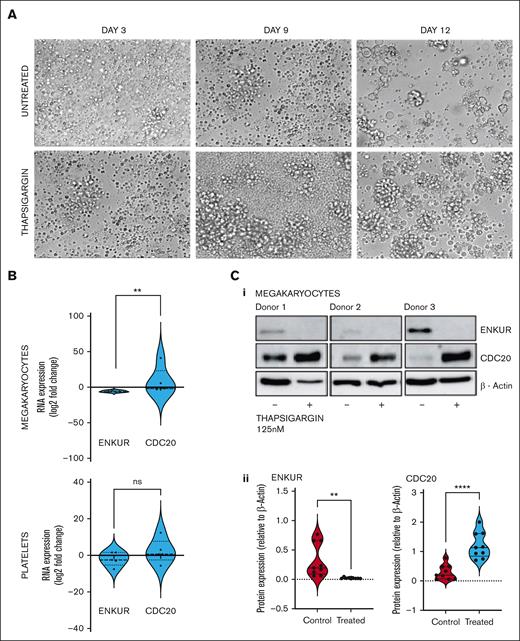
Effect of ER stress on CD34+–derived megakaryocytes
Considering that the differential and inversely correlated expression of ENKUR and CDC20 extends to the CD34+ cell–derived MKs and our hypothesis on the role of ER stress in this response, we assessed the RNA and protein levels of these markers after treatment of the cells with the sarco/endoplasmic reticulum Ca2+-ATPase inhibitor, thapsigargin (125 nM). CD34+ cells from peripheral blood of healthy donors were differentiated into megakaryocyte and platelet fractions, with thapsigargin introduced on day 7 of a 15-day culture (Table 1; Figure 5A; limited cell quantity of the platelet fraction enabled protein analyses in the megakaryocytic fraction alone).
Effect of ER stress on MKS derived from CD34+ cells from healthy donors. (A) Representative microscopic images of the CD34+ cells isolated from the peripheral blood of healthy donors over the 15-day culture period at original magnification ×20. The ER stressor, thapsigargin (125 nM), was added to the culture at day 7 and cultured for another 7 days. (B) RNA expression: cell culture from thapsigargin-treated CD34+-derived MKs (n = 7). (C) (1) Western blot of the thapsigargin-treated CD34+ cells–derived megakaryocytes (n = 3 in triplicate) and (2) densitometry for the blots. Densitometry was performed using the ImageJ software (∗∗∗∗P < .0001; ∗∗P < .01, when compared with untreated controls).
Effect of ER stress on MKS derived from CD34+ cells from healthy donors. (A) Representative microscopic images of the CD34+ cells isolated from the peripheral blood of healthy donors over the 15-day culture period at original magnification ×20. The ER stressor, thapsigargin (125 nM), was added to the culture at day 7 and cultured for another 7 days. (B) RNA expression: cell culture from thapsigargin-treated CD34+-derived MKs (n = 7). (C) (1) Western blot of the thapsigargin-treated CD34+ cells–derived megakaryocytes (n = 3 in triplicate) and (2) densitometry for the blots. Densitometry was performed using the ImageJ software (∗∗∗∗P < .0001; ∗∗P < .01, when compared with untreated controls).
Discussion
Here, we present the first report, to our knowledge, of enkurin (ENKUR) as a potential new peripheral biomarker and therapeutic strategy in MPNs. Our prior study20 profiling the platelet transcriptome in patients with chronic MPNs (n = 120) identified a progressive association in expression of several proteostasis-associated genes with the advancing disease subtype. Our findings were consistent with other studies demonstrating dysregulated proteostasis as a primary effector of myeloid transformation.18,19,55,56 The MPN platelet RNA-seq data also helped confirm the limited impact11 of treatment using JAK2-inhibitor RUX relative to the substantial disease burden in myelofibrosis and urged the need for the development of novel candidate drugs to be used alone or in combination with RUX for the treatment of MPNs.
Four candidate markers from the prior study (ENKUR, CDC20, CALR, and CREB3L1) were evaluated in this work based not only on their significant association with pathways related to proteostasis, cell proliferation, ER stress, or calcium signaling but also on their progressive increase or decrease in differential expression across the MPN chronic vs advanced subtypes (ET/PV to MF vs healthy donors) regardless of patient JAK2/CALR mutational status. Using CD34+ cells isolated from MPN patient bone marrow as well as healthy donor peripheral and cord blood, we generated ex vivo megakaryocyte and platelet fractions and discovered, at both RNA and protein levels, an inverse correlation between ENKUR, a calmodulin and transient receptor potential cation channel modulator26,29 (not previously associated with MPNs), and CDC20 (also termed Fizzy43), a cell cycle gene and an anaphase-promoting complex activator (with known associations with malignancies broadly42,57 and select studies in MPNs58). Lentiviral-mediated silencing of ENKUR was used to further confirm the negative association between ENKUR and CDC20, indicating a potential interaction via the PI3K-Akt pathway (Figure 6). Our data on the inverse association between enkurin, a membrane calcium influx adapter protein, and CDC20, a crucial cell cycle and proliferation regulator, point to a role for dysregulated calcium signaling in MPNs and the possibility for the ratio of expression between the 2 to be developed as a potential biomarker of MPN disease. The gradation, we note, in platelet expression of ENKUR/CDC20 across the subtypes, ET, PV, and MF, suggests also diverse cell cycle/differentiation states along a continuous spectrum of MPNs, in line with the clinical overlap of these disorders.
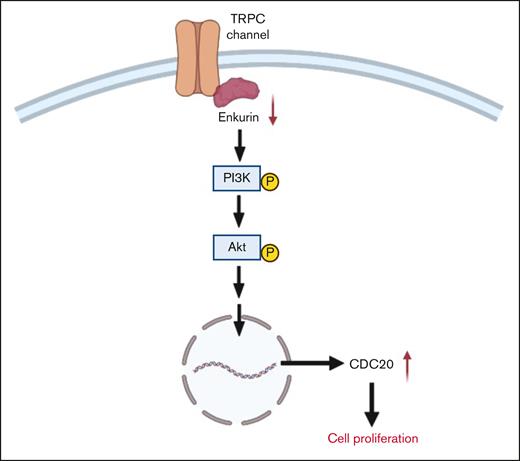
Probable mechanism for the role of ENKUR downregulation in cell proliferation. We hypothesize that, under normal conditions, enkurin at sufficient physiological levels is bound to PI3K via the SH3-binding domain, thus regulating its activation.26 However, in the setting of MPNs, downregulation of enkurin likely contributes to free PI3K and its prolonged activation, further activating Akt and potentially other signaling mechanisms, prompting overexpression of cell differentiation cycle genes, including CDC20,59 and cell proliferation.
Probable mechanism for the role of ENKUR downregulation in cell proliferation. We hypothesize that, under normal conditions, enkurin at sufficient physiological levels is bound to PI3K via the SH3-binding domain, thus regulating its activation.26 However, in the setting of MPNs, downregulation of enkurin likely contributes to free PI3K and its prolonged activation, further activating Akt and potentially other signaling mechanisms, prompting overexpression of cell differentiation cycle genes, including CDC20,59 and cell proliferation.
Next, in evaluating calreticulin (CALR), a well-established driver mutation in MPNs,31,60 as well as CREB3L1, an ER/UPR stress response transcription factor, high expression noted previously across patient platelet RNA-seq20 was confirmed in patient-derived specimens (platelet, whole blood, and bone marrow CD34+–cultured MKs) but only at the RNA (and not protein) levels. Future investigations applying genome editing to MPN models will be needed to ascertain the overlapping mechanisms of somatic alterations in CALR and their effect on function in altered calcium signaling and dysregulated ER protein folding61,62 in MPNs.
In summary, we offer 2 inversely associated MPN markers, ENKUR and CDC20, whose whole blood, platelet, and MK RNA and protein expression reflect MPN pathobiology irrespective of patient driver mutational status. These candidate markers may also expand our understanding of MPN pathology beyond the classical inflammatory signatures.63-66
Limitations of the study
There are several limitations to our study. Firstly, higher statistical power and longitudinal prospective data will be necessary to further confirm ENKUR/CDC20 as markers for MPN disease progression. Secondly, future investigations evaluating these signatures in patient-derived CD34+ cells may identify additional functional aspects of bone marrow and MPN pathology. Thirdly, we recognize that our data are not assessing mechanistic signaling of how loss of ENKUR expression or impaired calcium signaling contributes to myeloproliferation and will need to be specifically interrogated in MPN murine and other models. Follow-up studies evaluating the potential role of the PI3K/Akt pathway, in particular, the effect of pharmaceutical inhibitors already in use in MF will add significant value to the current study. Studies evaluating platelet function under impaired enkurin expression and calcium modulation are promising future directions. Above all, it will be important to assess how enkurin downregulation might be more broadly relevant to other hematological cancers, such as myelodysplastic syndromes and acute myeloid leukemia, or potentially unique to MPNs alone.
Acknowledgments
The authors thank the patients at the Stanford Cancer Center and the healthy donors at the Stanford Blood Center for their generous participation in this research, and the Canary Center at Stanford for Cancer Early Detection for access to flow cytometry and microscopy. The authors also thank Diwash Jangam for his assistance with the initial Paxgene PCR setup.
This work was funded by the MPN Research Foundation and the US National Institutes of Health grants 1K08HG010061-01A1 and 3UL1TR001085-04S1 (research re-entry award) (A.K.). Other support included 1S10OD018220 and 1S10OD021763 shared instrumentation grants to the Stanford Functional Genomics Facility and the Stanford Research Computing Center, and the Charles and Ann Johnson Foundation (J.G.).
Authorship
Contribution: A.K. conceived of the study and the team, secured funding, and wrote the manuscript; L.F. and J.G. provided samples and clinical annotation and reviewed the clinical data; A.K. and R.P. coordinated and oversaw sample acquisition and processing; S.M.S., Y.L., J.W., and K.M. performed the experiments and interpreted the analyses with A.K., R.P., H.M., J.G., and J.Z; all authors critically reviewed, edited, and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anandi Krishnan, Department of Pathology, Stanford University School of Medicine, Stanford, CA 94304; e-mail: anandi.krishnan@stanford.edu.
References
Author notes
∗S.M.S. and Y.L. contributed equally to this study.
†J.G. and J.Z. contributed equally to this study.
Data are available on request from the corresponding author, Anandi Krishnan (anandi.krishnan@stanford.edu).
The full-text version of this article contains a data supplement.