TO THE EDITOR:
Adults diagnosed with multiple myeloma (MM) carry an 11-fold higher risk of developing myelodysplastic syndrome and acute myelogenous leukemia (MDS/AML) than the general population.1,2 Standard risk factors for MDS/AML include genotoxic chemotherapy, radiation,3 and aging-induced expansion of hematopoietic clones harboring pathogenic mutations (ie, clonal hematopoiesis of indeterminate potential; CHIPs4). Although these risk factors may contribute, they are unlikely to be the exclusive drivers behind the increased incidence of MDS/AML among patients with MM.
Adults diagnosed with monoclonal gammopathy of undetermined significance (MGUS), another plasma cell dyscrasia that is a precursor to MM, have an eightfold increased risk of MDS/AML.2,5 The likelihood that a patient with MGUS develops MDS/AML is directly correlated with the concentration of monoclonal paraprotein in their blood serum.2 Although MM is treated with combinations of proteasome inhibitors, immunomodulatory drugs (IMiDs), and immunomodulatory agents followed by melphalan-conditioned autologous stem cell transplantation (ASCT6), MGUS is observed expectedly. That patients with MGUS nevertheless have an increased incidence of MDS/AML suggests a plasma cell–specific mechanism of leukemia initiation that is independent of genotoxic therapy.
CHIP expansion and evolution into MDS/AML is another potential explanation for the increased incidence of MDS/AML among patients with MM.4,7 In a recent study, Ghobrial et al sequenced autograft samples from patients with MM who were undergoing ASCT to determine whether autograft CHIPs affect posttransplantation clinical outcomes.5 Although this study found that autograft CHIPs were associated with overall poor posttransplantation clinical outcomes, CHIP-containing autograft cells did not transform into MDS/AML clones nor did they increase the likelihood of MM-associated MDS/AML. This study supports that CHIP expansion is unlikely to be the mechanism underlying the increased incidence of MDS/AML among patients with MM.
To assess another candidate mechanism, we evaluated whether patients with MM who subsequently develop MDS/AML have significant non-CHIP genetic lesions in their bone marrow before receiving MM-directed therapy. We identified 8 adults with MM who subsequently developed MDS/AML and for whom archived formalin-fixed and paraffin-embedded (FFPE) bone marrow and autograft samples were available. Clinical descriptors of these patients are included in Table 1, and the methodologies used to isolate, sequence, and analyze data are included in supplemental Materials. Institutional review board approval was granted at The Medical College of Wisconsin based on protocol #PRO00028417. The study was conducted in accordance with the declaration of Helsinki.
To detect the presence of genetic variants before ASCT and determine their temporal correlation with the development of MDS/AML, we performed whole exome sequencing of archived FFPE bone marrow samples obtained before ASCT (PRE), CD34+-selected autograft (AUTO), and FFPE bone marrow obtained at the time of MDS/AML diagnosis (POST). We were unable to obtain sufficient high-quality DNA or sequencing results from a subset of samples that are delineated within supplemental Table 1.
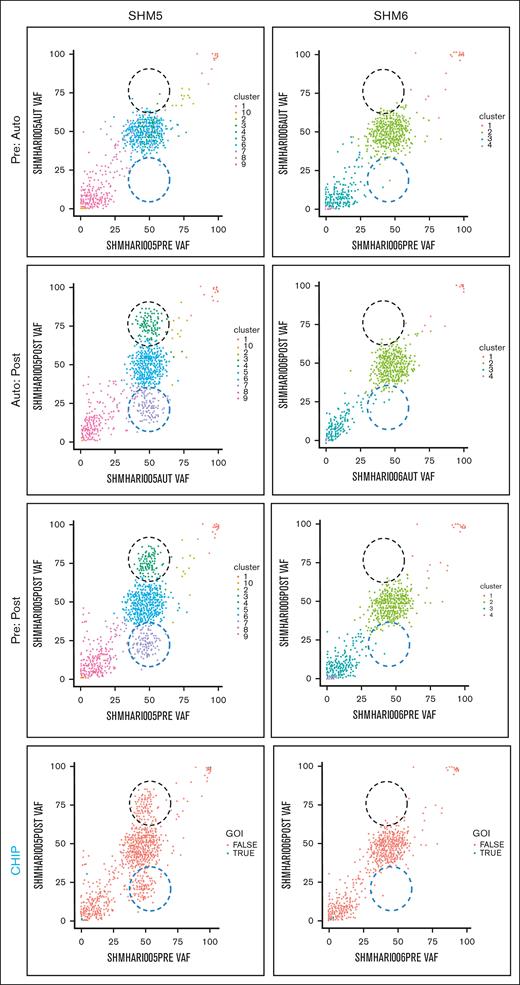
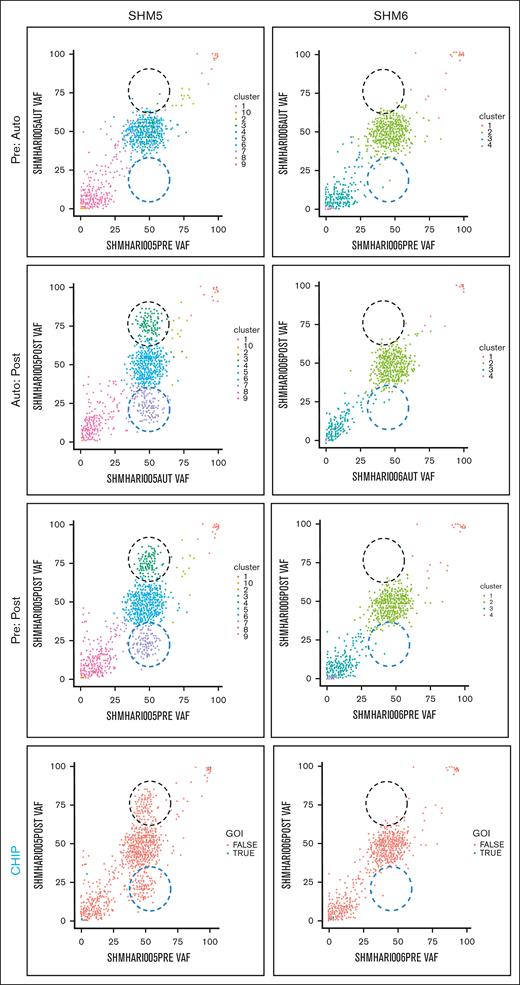
We explored the evolution of mutationally-defined clones by comparing variant allele frequencies (VAFs) in myeloid-malignancy associated genes between PRE, AUTO and POST samples (Figure 1;supplemental Materials). The complete catalog of variants organized based on timepoint and CHIP vs non-CHIP assignation are included in supplemental Materials. Only somatic lesions likely to be pathogenic were included in our analysis (supplemental Materials). Variants with VAF of 0.5 or 1 were excluded, because a nonhematopoietic tissue source that could differentiate germ line from somatic variants with those VAFs was not available. In 4 patients (SHM2, SHM4, SHM5, and SHM8), a subset of variants was enriched and a separate subset of variants was depleted between PRE and POST samples (black and blue dashed circles, respectively, in Figure 1). We did not detect variant enrichment from PRE to POST samples in either SHM3 or SHM6. Variants were subsequently categorized as CHIP (DNMT3A, TET2, ASXL1, TP53, JAK2, SF3B1, GNB1, CBL, SRSF2, and GNAS4) or non-CHIP genes (CHIP panels in Figure 1; supplemental Materials). Confirming Ghobrial’s earlier observation that CHIPs have a limited role in the development of MM-associated MDS/AML,5 the CHIP gene mutations detected in our patient cohort were minimal contributors to the subpopulation of enriched variants in POST samples. All gene mutations that increased over the time course or were present exclusively in the MDS/AML specimens are listed in supplemental Tables 2 and 3, respectively.
Comparison of PRE, AUTO, and POST VAFs in 2 patients with MM-associated MDS/AML. Graphical representation of VAFs between PRE:AUTO, AUTO:POST, and PRE:POST samples for SHM5 (variant enrichment) and SHM6 (no enrichment). The clinical timing of PRE is indicated in “MM biopsy” of Figure 1. Black dashed circle indicates variant enrichment. Blue dashed circle indicates variant reduction. Each dot indicates a single variant. In the panels labeled CHIP, blue points (ie, true) indicate CHIP gene variants. GOI, genes of interest (ie, CHIP genes).
Comparison of PRE, AUTO, and POST VAFs in 2 patients with MM-associated MDS/AML. Graphical representation of VAFs between PRE:AUTO, AUTO:POST, and PRE:POST samples for SHM5 (variant enrichment) and SHM6 (no enrichment). The clinical timing of PRE is indicated in “MM biopsy” of Figure 1. Black dashed circle indicates variant enrichment. Blue dashed circle indicates variant reduction. Each dot indicates a single variant. In the panels labeled CHIP, blue points (ie, true) indicate CHIP gene variants. GOI, genes of interest (ie, CHIP genes).
To determine the relative timing of variant enrichment, we compared VAFs between PRE and AUTO and between AUTO and POST samples. Despite a greater depth in sequencing of AUTO samples, there was no difference in variant enrichment between the PRE and AUTO samples from any of the 6 patients for whom all 3 timepoints could be sequenced. In the 4 patients for whom we found variant enrichment between PRE and POST samples (SHM2, SHM4, SHM5, and SHM8), these changes occurred between the AUTO and POST timepoints (Figure 1).
We set out to test the hypothesis that the genetic underpinnings of MDS/AML are present in the bone marrow of some patients with MM well before they undergo significant MM-directed therapy. Supporting this hypothesis, we detected non-CHIP genetic variants from PRE samples that were subsequently enriched at the time of MDS/AML diagnosis in 5 of our 8 patients (supplemental Table 2). The mechanism leading to the initial development of these variants remains unknown, but we speculate that immune dysregulation or other MM-induced bone marrow niche changes may contribute to it.
Our use of FFPE bone marrow samples precluded separation of MDS/AML cells from their background hematopoietic milieu, limiting our ability to definitively assign specific genetic variants to malignant myeloid cells vs hematopoietic stem cells, as has been done previously.8 A strength of our analysis strategy is the use of CD34+ enriched autograft samples for analysis, which is a clinically feasible strategy for larger clinical studies of MM-associated myeloid malignancies. Nonetheless, our data do not demonstrate the presence of a preexisting CHIP clone in the autograft that subsequently gives rise to MDS/AML in patients with MM and are consistent with findings reported by Shah et al.9 In the patients for whom we had PRE, POST, and AUTO samples and saw pre-existing variant enrichment at the time of MDS/AML diagnosis, the enrichment occurred after autograft collection. All 4 of these individuals had significant MM-directed therapy that included lenalidomide during the interval between ASCT and MDS/AML diagnosis. Thus, we cannot rule out a contribution of post-ASCT therapy with immunomodulatory drugs, such as lenalidomide to the evolution of variant-containing clones into MDS/AML in our patient cohort,5,10-13 a recently identified contributor to MDS/AML in other patients with MM.13 Comparison of VAFs between FFPE and CD34+ enriched autograft samples requires some caution; although we are reassured that in cases where all 3 samples were available, AUTO:POST (CD34+:FFPE) and PRE:POST (FFPE:FFPE) had similar trends for variant enrichment.
Future studies aimed at testing and validating our findings in an independent patient cohort will ideally incorporate a more sensitive, targeted approach along with consistent sample processing. As newer therapies prolong lives of adults with MM, the incidence of MDS/AML among adults with MM is likely to increase. We look forward to future studies to elicit the biological mechanisms that predispose adults with MM, and by inference MGUS, to develop pathogenic myeloid clones. These prospective investigations will need to explore early stages of MGUS/MM and the effects of MM and its bone marrow microenvironment on the evolution of myeloid malignancy.
Acknowledgments: Editorial assistance was provided by Patricia Walsh through the MCW Clinical and Translational Science Institute.
Support for this project was from the MCW Cancer Center and the Mellowes Center for Genomic Science and Precision Medicine (S.R. and P.H.), R03HL155174 (K.C.), K08HL127187 (K.C.), R01CA204231 (S.R.), P01HL149620 (S.R.), and the MCW Cancer Center Team Science Award (S.R., K.C., P.H., B.D., S.J., M.M., and J.M.K.).
Contribution: A.K., A.S., L.P., P.H., S.R., and K.C. designed the research; A.K., A.S., L.P., A.J.M., G.F.S., S.R., and K.C. implemented the studies detailed in this manuscript; S.R. and K.C. wrote the manuscript; and A.K., A.S., L.P., A.J.M., P.H., B.D., M.M., S.J., J.M.K., M.V.S., C.S., R.B., S.R., and K.C. reviewed, edited, and contributed key concepts to the article.
Conflict-of-interest disclosure: B.D. reports grant/research support from Amgen, Takeda, ACS, LLS, Janssen, Carsgen, Cartesian, Sanofi, Arcellx, Bristol Myers Squibb, and Genmab; is a member of the speaker's bureau in Celgene, Sanofi, and Karyopharm; and is on the advisory board of Amgen, Takeda, GSK, Janssen, AbbVie, Arcellx, Natera, Genentech, and Sanofi. P.H. reports employment in Iovance Biotherapeutics; receives honoraria from Sanofi, Amgen, Novartis, Bristol Myers Squibb, Janssen Oncology, and Karyopharm Therapeutics; has a consulting or advisory role in Sanofi, Takeda, Bristol Myers Squibb, Janssen, Amgen, Kite, a Gilead company, Karyopharm Therapeutics, and Pharmacyclics; and receives research funding from Millennium (Inst), Celgene (Inst), Onyx (Inst), and Spectrum Pharmaceuticals (Inst). M.V.S. reports research funding to institution from Astellas, Bristol Myers Squibb, MRKR Therapeutics, Celgene, and Marker Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Karen Carlson, Department of Medicine, Division of Hematology and Oncology, 9200 W Wisconsin Ave, Milwaukee, WI 53226; e-mail: kabarker@mcw.edu.
References
Author notes
∗S.R. and K.C. are joint senior authors
Data are available on request from the corresponding author, Karen Carlson (kabarker@mcw.edu).
Sequencing data cannot be made publicly available because they are patient-derived.
The full-text version of this article contains a data supplement.