Key Points
KLF1 directly drives expression of ZBTB7A, a key repressor of fetal γ-globin gene expression, in erythroid cells.
An erythroid-specific regulation mechanism allows upregulation of a novel ZBTB7A transcript in erythroid cells.
Abstract
Genes encoding the human β-like hemoglobin proteins undergo a developmental switch from fetal γ-globin to adult β-globin expression around the time of birth. β-hemoglobinopathies, such as sickle-cell disease and β-thalassemia, result from mutations affecting the adult β-globin gene. The only treatment options currently available carry significant adverse effects. Analyses of heritable variations in fetal hemoglobin (HbF) levels have provided evidence that reactivation of the silenced fetal γ-globin genes in adult erythroid cells is a promising therapy. The γ-globin repressor BCL11A has become the major focus, with several studies investigating its regulation and function as a first step to inhibiting its expression or activity. However, a second repression mechanism was recently shown to be mediated by the transcription factor ZBTB7A/LRF, suggesting that understanding the regulation of ZBTB7A may also be useful. Here we show that Krüppel-like factor 1 (KLF1) directly drives expression of ZBTB7A in erythroid cells by binding to its proximal promoter. We have also uncovered an erythroid-specific regulation mechanism, leading to the upregulation of a novel ZBTB7A transcript in the erythroid compartment. The demonstration that ZBTB7A, like BCL11A, is a KLF1 target gene also fits with the observation that reduced KLF1 expression or activity is associated with HbF derepression.
Introduction
Diseases such as sickle-cell anemia and β-thalassemia, caused by the disruption of the adult β-globin gene and known collectively as β-globinopathies, are among the most common genetic diseases globally. Because only the adult β-globin gene is typically disrupted in these diseases, reactivation of fetal γ-globin expression in adult patients poses an attractive therapeutic option.1,2 Therefore, identification and investigation of factors involved in silencing fetal globin may inform useful therapeutic strategies.
Since its discovery, BCL11A has been recognized as the central factor in globin switching.3-6 Recently, the transcriptional repressor ZBTB7A has been identified as a BCL11A-independent silencer of γ-globin.7 Inactivation of ZBTB7A derepressed γ-globin expression in a humanized transgenic mouse model, as well as in human cells, and ZBTB7A normally represses expression of the γ-globin genes.7 ZBTB7A represses γ-globin expression by recruiting a unique NuRD repressor complex, independently of BCL11A. This establishes ZBTB7A as a new molecule to explore with respect to treating β-hemoglobinopathies. In particular, understanding its regulation may suggest avenues for altering its expression in erythroid cells and may inform our understanding of its role in the transcriptional network driven by master erythroid transcription factors such as Krüppel-like factor 1 (KLF1) and GATA-1. KLF1 itself is of considerable interest, because heterozygosity of KLF1 and reduction of KLF1 function have been associated with reduced levels of the repressor BCL11A and derepression of the fetal globin genes.8,9 Here we demonstrate that KLF1 also drives the expression of the second potent γ-globin repressor, ZBTB7A.
Methods
Klf1−/− mice
Ethics approval was obtained from the Animal Care and Ethics Committee, University of New South Wales, approval no. 09/128A. This line has been described previously.10
Cell culture
Culture conditions for KLF1-inducible estrogen receptor (K1ER) erythroblast cells, human umbilical cord blood–derived erythroid progenitor-2 (HUDEP-2) cells, and human CD34+ cord blood cells are described in the supplemental Methods.
Western blotting
Electrophoretic mobility shift assay
COS cells were transfected with 5 μg of pMT3-KLF1, pMT3-GATA-1, or pMT3 empty using FuGENE6 (Roche Diagnostics) as instructed by the supplier. Electrophoretic mobility shift assays were performed as described previously14 (probes in supplemental Methods). α-mKLF114 and α-GATA-1 (clone N6, sc-265; Santa Cruz Biotechnology) antibodies were used to confirm the identity of the complexes.
RNA analyses
RNA was prepared as previously described.15 Details of quantitative real-time reverse transcription polymerase chain reaction and 5′ rapid amplification of complementary DNA ends (RACE) can be found in the supplemental Methods.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed as previously described16 on 2 replicates of induced K1ER cells and HUDEP-2 cells.16 α-KLF1 (PIEPA5-18031; Thermo Scientific) and anti-goat immunoglobulin G (SCZSC-2028; Santa Cruz Biotechnology) antibodies were used. High-throughput sequencing and bioinformatics are detailed in the supplemental Methods.
Results
We initially examined Zbtb7a transcript levels in Klf1−/− fetal livers and observed a reduction in expression compared with wild-type controls (Figure 1A-B). Levels of ZBTB7A protein were also significantly lower in Klf1−/− fetal livers at E14.5 (Figure 1C; supplemental Figure 1A). This is consistent with preliminary microarray studies from our group and others showing a downregulation of Zbtb7a in the absence of KLF1.15,17
Zbtb7a is downregulated in the absence of KLF1. Transcript levels of Zbtb7a were determined by quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) in Klf1+/+ and Klf1−/− fetal livers at E13.5 (A) and E14.5 (B). In each instance, Klf1−/− samples were set to 1 (n = 5 per genotype). (C) Representative western blot of ZBTB7A expression in nuclear extracts isolated from fetal liver at E14.5. β-actin is presented as a loading control. (D) Transcript levels of Zbtb7a were determined by quantitative real-time RT-PCR in K1ER cells induced with tamoxifen for 0, 2, 4, 6, 8, 24, and 48 hours. The 0 hour time point was set to 1 (n = 2 per condition). (E) Representative western blot showing KLF1 and ZBTB7A expression in nuclear extracts isolated from K1ER cells at various time points postinduction. β-actin is presented as a loading control. (F) Transcript levels of Zbtb7a were determined by quantitative real-time RT-PCR in K1ER cells induced with tamoxifen for 0, 0.25, 0.5, 1, 2, 3, 4, and 48 hours. The 0 hour time point was set to 1 (n = 4). (G) K1ER cells were treated with either cycloheximide (CHX), tamoxifen, or a combination of both and harvested at 0, 2, 4, and 6 hours posttreatment. CHX was added to the appropriate cells 30 minutes before commencement induction with tamoxifen (n = 4 for each treatment). All RT-PCR values were normalized to 18S ribosomal RNA. Error bars represent the standard error of the mean. *P < .05; **P < .01 (paired Student 2-tailed t test).
Zbtb7a is downregulated in the absence of KLF1. Transcript levels of Zbtb7a were determined by quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) in Klf1+/+ and Klf1−/− fetal livers at E13.5 (A) and E14.5 (B). In each instance, Klf1−/− samples were set to 1 (n = 5 per genotype). (C) Representative western blot of ZBTB7A expression in nuclear extracts isolated from fetal liver at E14.5. β-actin is presented as a loading control. (D) Transcript levels of Zbtb7a were determined by quantitative real-time RT-PCR in K1ER cells induced with tamoxifen for 0, 2, 4, 6, 8, 24, and 48 hours. The 0 hour time point was set to 1 (n = 2 per condition). (E) Representative western blot showing KLF1 and ZBTB7A expression in nuclear extracts isolated from K1ER cells at various time points postinduction. β-actin is presented as a loading control. (F) Transcript levels of Zbtb7a were determined by quantitative real-time RT-PCR in K1ER cells induced with tamoxifen for 0, 0.25, 0.5, 1, 2, 3, 4, and 48 hours. The 0 hour time point was set to 1 (n = 4). (G) K1ER cells were treated with either cycloheximide (CHX), tamoxifen, or a combination of both and harvested at 0, 2, 4, and 6 hours posttreatment. CHX was added to the appropriate cells 30 minutes before commencement induction with tamoxifen (n = 4 for each treatment). All RT-PCR values were normalized to 18S ribosomal RNA. Error bars represent the standard error of the mean. *P < .05; **P < .01 (paired Student 2-tailed t test).
We employed the KLF1-inducible cell line K1ER18 to investigate the dynamics of this regulation. Zbtb7a transcript and protein levels were examined postinduction of KLF1 (Figure 1D-E). Zbtb7a transcript levels had increased rapidly to sixfold above basal levels by 2 hours postinduction and dropped to a twofold increase by 4 hours, which was maintained until the final time point of 48 hours. ZBTB7A protein levels increased at 6 hours, continuing throughout the time course to 48 hours postinduction (Figure 1E; supplemental Figure 1B). We next performed a shorter time course to determine how quickly Zbtb7a is upregulated in response to KLF1 and found an increase as early as 30 minutes postinduction that peaked at 1 to 3 hours (Figure 1F). The rapid response of Zbtb7a levels to induction of KLF1 prompted us to test whether Zbtb7a was directly activated by KLF1.
The KLF1-ER protein is constitutively expressed in K1ER cells and sequestered in an inactive state in the absence of tamoxifen. This enabled us to determine whether KLF1 directly activates transcription of Zbtb7a using the translation inhibitor cycloheximide (CHX). K1ER cells were exposed to CHX 30 minutes before induction with tamoxifen. In the presence of CHX, Zbtb7a transcripts were upregulated (Figure 1G) to levels similar to those observed previously during induction without CHX. This suggests KLF1 directly activates Zbtb7a, because de novo protein synthesis was not required for its upregulation.
We next performed ChIP sequencing (ChIP-Seq) experiments in both mouse and human cell lines to determine whether KLF1 binds directly to the ZBTB7A promoter. Mouse ChIP-Seq experiments were performed in K1ER cells induced with tamoxifen for 2 hours (Figure 2; supplemental Figure 2A-B). Human ChIP-Seq experiments were performed in HUDEP-2 cells19 (Figure 3; supplemental Figure 2C-D), which are at the basophilic stage and which we have found to have levels of KLF1 expression similar to those of CD34+ cells at week 3 of differentiation. In both mouse and human contexts, we identified the ZBTB7A promoter as one of the most highly enriched KLF1 peaks (Figures 2D-E and 3D-E, respectively). Our data represent, to our knowledge, the first KLF1 ChIP-Seq analysis in HUDEP-2 cells.
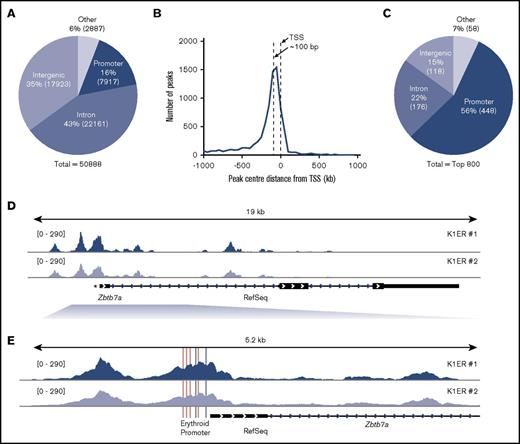
K1ER KLF1 ChIP-Seq analysis. (A) Distribution of all KLF1 peaks. Promoters are defined as the region from −1000 bp to +1000 bp around the transcription start site (TSS) of RefSeq genes. Peaks that fell into CDS exons 4′ and 3′ untranslated region exons and transcription termination sites (−100 bp to +1 kb) were all labeled as other. Percentages lying in each region are given, and absolute peak numbers are shown in parentheses. (B) Histogram of distance of promoter peak centers to the corresponding RefSeq TSS with 50-bp bins. (C) The top 800 KLF1 peaks within different genomic regions. (D) KLF1 enrichment across the gene body of Zbtb7a. (E) KLF1 enrichment at the promoter region of Zbtb7a, showing the conserved CACCC and GATA consensus sequences in pink and gray, respectively.
K1ER KLF1 ChIP-Seq analysis. (A) Distribution of all KLF1 peaks. Promoters are defined as the region from −1000 bp to +1000 bp around the transcription start site (TSS) of RefSeq genes. Peaks that fell into CDS exons 4′ and 3′ untranslated region exons and transcription termination sites (−100 bp to +1 kb) were all labeled as other. Percentages lying in each region are given, and absolute peak numbers are shown in parentheses. (B) Histogram of distance of promoter peak centers to the corresponding RefSeq TSS with 50-bp bins. (C) The top 800 KLF1 peaks within different genomic regions. (D) KLF1 enrichment across the gene body of Zbtb7a. (E) KLF1 enrichment at the promoter region of Zbtb7a, showing the conserved CACCC and GATA consensus sequences in pink and gray, respectively.
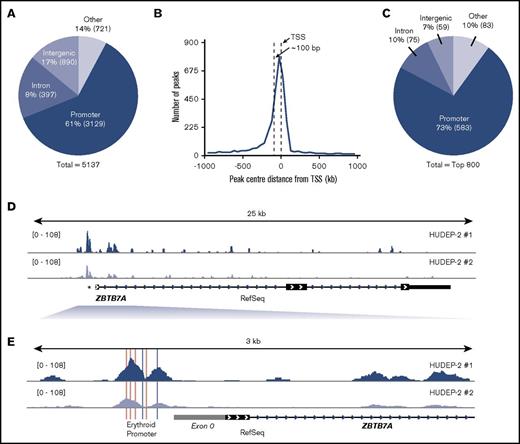
HUDEP-2 KLF1 ChIP-Seq analysis. (A) Distribution of all KLF1 peaks. Promoters are defined as the region from −1000 bp to +1000 bp around the TSS of RefSeq genes. Peaks that fell into CDS exons 4′ and 3′ UTR exons and transcription termination sites (−100 bp to +1 kb) were all labeled as other. Percentages lying in each region are given, and absolute peak numbers are shown in parentheses. (B) Histogram of distance of promoter peak centers to the corresponding RefSeq TSS with 50-bp bins. (C) The top 800 KLF1 peaks within different genomic regions. (D) KLF1 enrichment across the gene body of ZBTB7A. (E) KLF1 enrichment at the promoter region of ZBTB7A, showing the conserved CACCC and GATA consensus sequences in pink and gray, respectively, as well as the placement of exon 0 in gray.
HUDEP-2 KLF1 ChIP-Seq analysis. (A) Distribution of all KLF1 peaks. Promoters are defined as the region from −1000 bp to +1000 bp around the TSS of RefSeq genes. Peaks that fell into CDS exons 4′ and 3′ UTR exons and transcription termination sites (−100 bp to +1 kb) were all labeled as other. Percentages lying in each region are given, and absolute peak numbers are shown in parentheses. (B) Histogram of distance of promoter peak centers to the corresponding RefSeq TSS with 50-bp bins. (C) The top 800 KLF1 peaks within different genomic regions. (D) KLF1 enrichment across the gene body of ZBTB7A. (E) KLF1 enrichment at the promoter region of ZBTB7A, showing the conserved CACCC and GATA consensus sequences in pink and gray, respectively, as well as the placement of exon 0 in gray.
The distribution of KLF1 peaks across promoters, exons, introns, and other regions across the Mus musculus (mm10/NCBI38) and Homo sapiens (hg19/NCBI38) genomes were analyzed based on RefSeq annotations as previously reported.20 Briefly, promoter regions were defined as being −10 to +1 kb from the RefSeq TSS, intronic regions were those lying between exons, and intergenic regions made up the remainder of the genome. Peaks that fell into coding exons or 5′ and 3′ untranslated region exons or close to the transcription termination sites (−100 bp to +1 kb) were labeled as other. A total of 50 888 KLF1 peaks for K1ER cells (Figure 2A) and 5137 for HUDEP-2 cells (Figure 3A) were identified as overlapping between the 2 replicate sets of each cell type.
As previously reported,21 a large number of KLF1 peaks fell within introns, with 43% in K1ER cells and 17% in HUDEP-2 cells (Figures 2A and 3A), which is highly enriched above their representation in the genome. An enrichment of peaks was also found at promoter regions, with 16% in K1ER cells and 61% in HUDEP-2 cells. A large representation of intergenic regions was also seen, with 35% in K1ER cells and 14% in HUDEP-2 cells; this was expected, because these regions make up the majority of the genome. The precise location of the promoter peaks relative to the TSS is shown in Figures 2B and 3B. The confluence of peak centers was located approximately 100 bp upstream from the TSS. Next, we tested whether our ChIP-Seq data set included peaks at well-characterized KLF1 target genes. We looked first at Hbb-b110 (supplemental Figure 2A,C) and Klf322 (supplemental Figure 2B,D). KLF1 is enriched at the promoters (indicated by *) of both genes.
The addition of the ER tag in K1ER cells may affect the binding of KLF1, which could explain the skewed distribution of the pie chart in supplemental Figure 2A toward intergenic regions. We therefore chose to focus our findings on the top 800 peaks (Figure 2C). We believe these best represent wild-type KLF1 binding sites, because the addition of the tag is more likely to disrupt binding of weaker sites rather than facilitate novel binding sites. We included an analysis of the top 800 peaks in HUDEP-2 cells as a direct comparison (Figure 3C). Here, much greater KLF1 enrichment was found at promoters, with 56% in K1ER cells and 73% in HUDEP-2 cells, suggesting that KLF1 predominantly binds strongly to promoters. Intronic regions were still enriched compared with their representation in the genome, with 22% in K1ER cells and 10% in HUDEP-2 cells, whereas intergenic binding was far less abundant, with 15% in K1ER cells and 7% in HUDEP-2 cells.
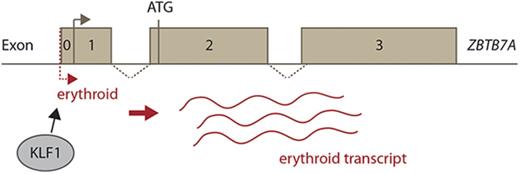
Many important erythroid genes have erythroid-specific transcripts, allowing for high-level expression in the erythroid compartment. Considering the important role of ZBTB7A in erythroid cells, we hypothesized that it may also be alternatively regulated in these cells.
We investigated the expression data from the FANTOM5 project.23,24 Here we found evidence of an erythroid transcript, of currently unknown significance or function, highly expressed in reticulocytes (supplemental Figure 3A). To further examine this, we employed 5′ RACE, using a reverse primer specific for exon 2 of ZBTB7A. RNA was extracted from human hematopoietic stem and progenitor cells (HSPCs) isolated from cord blood and differentiated toward the erythroid lineage as previously described.25 At differentiation week 1, these cells resemble HSPCs, and at week 3, they mainly consist of reticulocytes. 5′ RACE revealed the presence of a novel ZBTB7A transcript expressed in differentiated cells (Figure 4A). Sequencing analysis revealed that this transcript has a transcriptional start site slightly upstream of the broadly expressed ZBTB7A transcript, supporting the data from FANTOM5 and indicating that no alternative splicing occurs with exon 2 (Figure 3E; supplemental Figure 3B-C).
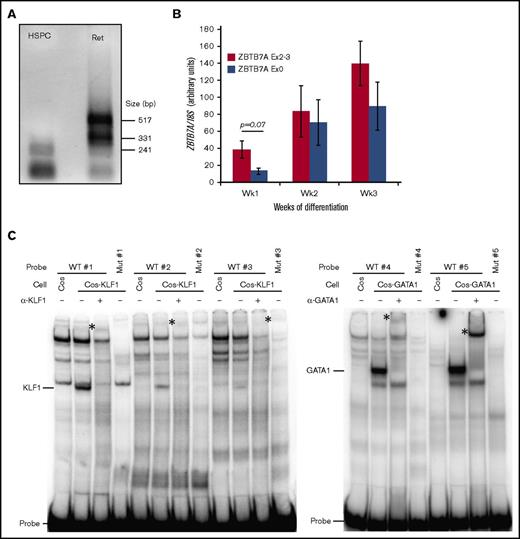
Erythroid-specific promoter activates a novel ZBTB7A transcript. (A) Messenger RNA from CD34+ hematopoietic stem cells differentiated toward the erythroid lineage for 1 (HSPC) and 3 (Ret) weeks was subjected to 5′ RACE using a reverse primer specific for exon 2 of ZBTB7A and analyzed by agarose gel electrophoresis. The larger bands present in the reticulocyte samples were sequenced and revealed a novel transcriptional start site upstream of the previously annotated ZBTB7A RefSeq transcriptional start site. (B) Polymerase chain reaction quantification revealing that transcripts containing exon 0 are lowly expressed in HSPC cells but highly upregulated during differentiation (n = 3 per time point). Values were normalized to 18S ribosomal RNA levels. Error bars represent the standard error of the mean (P = .068; paired Student 2-tailed t test). (C) Nuclear extracts were harvested from COS cells and COS cells overexpressing either KLF1 or GATA-1. Binding of these proteins to wild-type (WT) and mutant (Mut) KLF1 and GATA-1 consensus sequences of the ZBTB7A promoter were analyzed by electrophoretic mobility shift assay using radiolabeled probes. Free-probe, KLF1:DNA, and GATA-1:DNA complexes are as indicated. The identities of these complexes were confirmed by supershifting (*) with antibodies specific for KLF1 and GATA-1.
Erythroid-specific promoter activates a novel ZBTB7A transcript. (A) Messenger RNA from CD34+ hematopoietic stem cells differentiated toward the erythroid lineage for 1 (HSPC) and 3 (Ret) weeks was subjected to 5′ RACE using a reverse primer specific for exon 2 of ZBTB7A and analyzed by agarose gel electrophoresis. The larger bands present in the reticulocyte samples were sequenced and revealed a novel transcriptional start site upstream of the previously annotated ZBTB7A RefSeq transcriptional start site. (B) Polymerase chain reaction quantification revealing that transcripts containing exon 0 are lowly expressed in HSPC cells but highly upregulated during differentiation (n = 3 per time point). Values were normalized to 18S ribosomal RNA levels. Error bars represent the standard error of the mean (P = .068; paired Student 2-tailed t test). (C) Nuclear extracts were harvested from COS cells and COS cells overexpressing either KLF1 or GATA-1. Binding of these proteins to wild-type (WT) and mutant (Mut) KLF1 and GATA-1 consensus sequences of the ZBTB7A promoter were analyzed by electrophoretic mobility shift assay using radiolabeled probes. Free-probe, KLF1:DNA, and GATA-1:DNA complexes are as indicated. The identities of these complexes were confirmed by supershifting (*) with antibodies specific for KLF1 and GATA-1.
To validate this novel transcript in erythroid differentiation, we performed quantitative polymerase chain reaction in human HSPCs, differentiated toward the erythroid lineage as described.25 Primers specific for the extended first exon, termed exon 0, were used to investigate the erythroid transcript, whereas primers spanning the exon 2 to 3 boundary were used to ascertain total ZBTB7A levels. We found that exon 0 was not highly expressed in cells at week 1 (HSPCs), with the difference in expression between constitutive ZBTB7A and exon 0 approaching statistical significance (P = .068). Expression of this transcript, however, increased during erythroid differentiation at weeks 2 and 3 (Figure 4B). In fact, this transcript seemed to make up the majority of total ZBTB7A transcripts at these later stages of differentiation.
The promoter region of ZBTB7A contains KLF1 (CACCC boxes) and GATA-1 (GATA boxes) binding sites that are conserved between human and mouse (Figure 3C), indicated by the pink and gray bars, respectively (Figures 2E and 3E). These sites may be instrumental in generating the erythroid transcript. Our ChIP-Seq analysis revealed KLF1 binds over this region in human and mouse cells (Figures 2E and 3E). We also demonstrated that KLF1 and GATA-1 can bind to these sites in vitro and that mutations in these sites abolish binding (Figure 4C). These sites may represent an erythroid-specific promoter for the regulation of the novel transcript highly expressed in erythroid cells.
Discussion
Here we have demonstrated that KLF1 directly activates ZBTB7A by binding to its proximal promoter in both human and mouse erythroid cells. We have also uncovered an erythroid-specific regulation mechanism, leading to the upregulation of a novel ZBTB7A transcript in the erythroid compartment.
Although we have demonstrated that Zbtb7a is directly upregulated by KLF1 in murine erythroid cells, it is interesting to note that this upregulation seems to be relatively modest. Zbtb7a is still detectable in both Klf1−/− fetal liver cells and uninduced K1ER cells, indicating that other transcription factors may also play a part in Zbtb7a activation. Previous research has demonstrated that GATA-1 activates Zbtb7a.26 Because GATA-1 is known to activate the expression of Klf1,27 it is conceivable that both KLF1 and GATA-1 act together to upregulate Zbtb7a, as they do at many erythroid genes.21
Our ChIP-Seq results demonstrate that KLF1 binding is highly enriched at the promoter of ZBTB7A in both murine and human cell lines. There is a large difference in the total number of peaks between these 2 cell lines. Because the K1ER cells are a KLF1 overexpression cell line, and HUDEP-2 cells are much closer to primary cells, this is to be expected. As stated under Results, the addition of the ER tag to KLF1 in K1ER cells may also contribute to this difference. It should be noted, however, that previous analyses using these cells found targets similar to those seen in Klf1−/− cells, indicating that the addition of the ER tag has minimal effects on KLF1 function and binding.
We also uncovered the upregulation of a novel transcript in erythroid cells. This transcript is an extension of the first exon; however, because the translation start site lies in exon 2, this does not give rise to a novel protein. This mirrors what is seen with other erythroid targets, such as Klf3.22 Here, KLF1 upregulates an erythroid-specific first exon, termed Klf3 exon 1b, which is alternatively spliced to exon 2. As is the case with ZBTB7A, this alternative transcript does not produce a novel protein, because the start site of translation of Klf3 also lies in exon 2. Although the exact function of both Klf3 and Zbtb7a KLF1-dependent erythroid transcripts are hereto unknown, we hypothesize that their role is to provide a mechanism for the specific upregulation of these proteins in erythroid cells.
In the case of Zbtb7a, it is interesting to note that the data from FANTOM5 suggest this transcript is also highly expressed in monocytes and mast cells. Both GATA-1 and KLF1 have been shown to play a role in mast cells,28,29 and GATA-1 has also been demonstrated to be expressed in monocytes,30 which may account for the expression of this transcript in these cells.
Given the importance of ZBTB7A in the process of globin switching, understanding how KLF1 and GATA-1 cooperate to regulate its expression in an erythroid context may open avenues for modulating this expression and inform therapeutic strategies for β-globinopathies. The finding that first BCL11A and now ZBTB7A are direct KLF1 target genes is of interest given that reduced KLF1 levels are also associated with fetal globin derepression. Our results suggest that the derepression may be attributable to reduced ZBTB7A as well as to the previously demonstrated reduction in BCL11A.8
The ChIP-Seq data from this article are available from the Gene Expression Omnibus under accession number GSE97671.
The full-text version of this article contains a data supplement.
Acknowledgments
The ChIP-Seq samples were sequenced at the Ramaciotti Centre for Genomics at the University of New South Wales (UNSW), Sydney, Australia.
This work was supported by funding from the Australian National Health and Medical Research Council (APP1098391) (M.C.), a UNSW Australian Postgraduate Award (L.J.N.), and a UNSW University International Postgraduate Award (B.W.).
Authorship
Contribution: L.J.N. performed research, analyzed data, and wrote the manuscript; J.B. analyzed data; B.W. performed research; A.P.W.F., S.P., and R.C.M.P. designed research; R.K. and Y.N. contributed vital new reagents (HUDEP-2 cells); K.G.R.Q. and M.C. designed research and wrote the manuscript; and all authors reviewed and contributed to the final manuscript and approved the manuscript and data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Merlin Crossley, School of Biotechnology and Biomolecular Sciences, University of New South Wales, NSW 2052, Australia; e-mail: m.crossley@unsw.edu.au.