TO THE EDITOR:
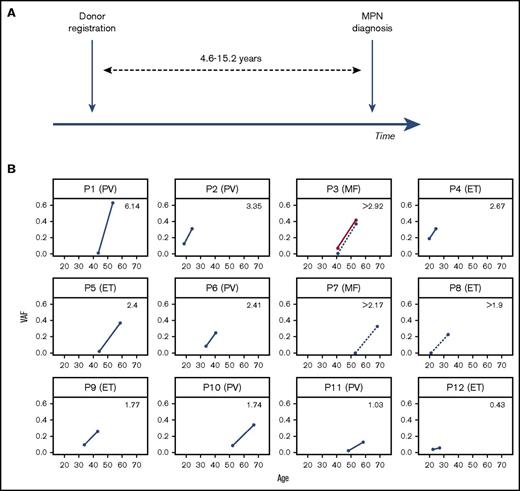
The JAK2 V617F mutation is the most common somatic mutation in the classical myeloproliferative neoplasms (MPNs), present in >95% of cases of polycythemia vera (PV) and ∼50% of essential thrombocythemia (ET) and myelofibrosis (MF).1-4 It is usually the sole identifiable driver mutation in MPNs5 and was recently also identified as a driver of age-related clonal hemopoiesis in healthy individuals.6-9 In order to investigate the preclinical clonal evolution of MPNs, we identified 12 individuals with a JAK2 V617F mutant MPN, who 4.6 to 15.2 years previously (median 10.2 years) had also donated blood to register with the Cyprus Bone Marrow Donor Registry at the Karaiskakio Foundation (Table 1; Figure 1A).
Participant characteristics
| Patient ID . | Age at MPN diagnosis (y) . | Sex . | Diagnosis . | WBC count (×106/L) . | Hb (g/L) . | Platelet count (×106/L) . | Time between samples (y) . |
|---|---|---|---|---|---|---|---|
| P1 | 53.6 | F | PV | 17.3 | 194 | 550 | 10.1 |
| P2 | 24.2 | F | PV | n/a | n/a | n/a | 5.5 |
| P3 | 53.4 | F | MF | 4.2 | 112 | 130 | 12.6 |
| P4 | 24.5 | M | ET | 8.4 | 15.6 | 600 | 4.6 |
| P5 | 58.7 | F | ET | 8.94 | 142 | 637 | 14.5 |
| P6 | 40.6 | F | PV | 7.7 | 168 | 602 | 6.8 |
| P7 | 68.2 | M | MF | 8.1 | 81 | 74 | 15.2 |
| P8 | 33 | M | ET | 10.3 | 149 | 776 | 12.1 |
| P9 | 43.2 | M | ET | 12.1 | 148 | 793 | 9.4 |
| P10 | 66.9 | F | PV | 7.6 | 154 | 830 | 14.7 |
| P11 | 58.4 | M | PV | 12.7 | 220 | 423 | 10.3 |
| P12 | 27 | F | ET | 5.1 | 135 | 750 | 4.6 |
| Patient ID . | Age at MPN diagnosis (y) . | Sex . | Diagnosis . | WBC count (×106/L) . | Hb (g/L) . | Platelet count (×106/L) . | Time between samples (y) . |
|---|---|---|---|---|---|---|---|
| P1 | 53.6 | F | PV | 17.3 | 194 | 550 | 10.1 |
| P2 | 24.2 | F | PV | n/a | n/a | n/a | 5.5 |
| P3 | 53.4 | F | MF | 4.2 | 112 | 130 | 12.6 |
| P4 | 24.5 | M | ET | 8.4 | 15.6 | 600 | 4.6 |
| P5 | 58.7 | F | ET | 8.94 | 142 | 637 | 14.5 |
| P6 | 40.6 | F | PV | 7.7 | 168 | 602 | 6.8 |
| P7 | 68.2 | M | MF | 8.1 | 81 | 74 | 15.2 |
| P8 | 33 | M | ET | 10.3 | 149 | 776 | 12.1 |
| P9 | 43.2 | M | ET | 12.1 | 148 | 793 | 9.4 |
| P10 | 66.9 | F | PV | 7.6 | 154 | 830 | 14.7 |
| P11 | 58.4 | M | PV | 12.7 | 220 | 423 | 10.3 |
| P12 | 27 | F | ET | 5.1 | 135 | 750 | 4.6 |
F, female; Hb, hemoglobin concentration; M, male; MF, idiopathic myelofibrosis; n/a, not available; WBC, white blood cell.
Preclinical expansion of JAK2 V617F clones in 12 MPN patients. (A) Schema of blood sample collection from 12 individuals at the time of registration as stem cell donors at the Cyprus Bone Marrow Donor Registry and at the time they were diagnosed with MPN 4.6 to 15.2 years later. (B) Variant allele fraction (VAF) sizes of JAK2 V617F–positive clones at the 2 time points against the age of participants at the time. The specific diagnosis is indicated in brackets next to each patient’s ID, and the average annual rise in JAK2 V617F VAF is indicated in the upper right quadrant of each plot. Samples P3, P7, and P8 had no detectable JAK2 V617F at donor registration. The VAF rise for SRSF2 P95R in patient P3 is show in red.
Preclinical expansion of JAK2 V617F clones in 12 MPN patients. (A) Schema of blood sample collection from 12 individuals at the time of registration as stem cell donors at the Cyprus Bone Marrow Donor Registry and at the time they were diagnosed with MPN 4.6 to 15.2 years later. (B) Variant allele fraction (VAF) sizes of JAK2 V617F–positive clones at the 2 time points against the age of participants at the time. The specific diagnosis is indicated in brackets next to each patient’s ID, and the average annual rise in JAK2 V617F VAF is indicated in the upper right quadrant of each plot. Samples P3, P7, and P8 had no detectable JAK2 V617F at donor registration. The VAF rise for SRSF2 P95R in patient P3 is show in red.
First, we interrogated all 24 samples for 15 myeloid mutation hot spots including JAK2 V617 (supplemental Table 1), using a previously described multiplex polymerase chain reaction/MiSeq sequencing protocol that reliably detects nucleotide substitutions present at a VAF ≥0.008.6 Additionally, for 12 samples with sufficient DNA available, we performed targeted DNA capture for all exons of 41 genes recurrently mutated in myeloid neoplasms (supplemental Table 2) using targeted capture with custom RNA baits (SureSelect, Agilent Technologies ELID ID: 0735431) followed by sequencing on Illumina HiSeq 2500. Substitutions and indels were identified and quantified using Mutation Identification and Analysis Software as described.6,10 Finally, we genotyped archival registry samples for rs12343867, a single nucleotide polymorphism (C/T) linked to the JAK2 46/1 haplotype and polymorphisms at the TERT, SH2B3, TET2, MECOM, and MYB genes that are associated with a predisposition to MPN.11,12 The study was approved by the Cyprus National Bioethics Committee (EEBK/EΠ/2014/11) and performed in accordance with the Declaration of Helsinki.
Amplicon sequencing returned a median coverage of 6641 reads per nucleotide at the studied hot spots (excluding NPM1 exon 12). This confirmed the presence of JAK2 V617F in all 12 diagnostic and 9 of 12 archival samples (supplemental Table 3). The remaining 3 samples were JAK2 V617F negative at the sensitivity of our assay (VAF ≥0.008). The only other hot spot mutation identified was SRSF2 P95R in patient P3 (see later). Pull-down sequencing of all exons of 41 genes from 12 samples with sufficient DNA returned an average coverage of 1978 reads per nucleotide and showed a close correlation in VAF quantitation for both JAK2 V617F and SRSF2 P95R with amplicon sequencing (supplemental Table 3).
The JAK2 V617F VAF at MPN diagnosis differed between patients as expected13 ; however, the average rate of clonal growth also varied widely, ranging from 0.36% to 6.2% per annum (Figure 1B). Targeted exon capture from 12 of 24 samples only identified 1 co-mutation with a VAF >0.02, SRSF2 P95R in patient P3 who had a diagnosis of MF (supplemental Table 3). As this locus was also studied by amplicon sequencing, we were able to quantify the SRSF2 P95R VAF in both the diagnostic and the archival sample taken 12.6 years earlier. The MPN diagnostic sample VAFs for JAK2 V617F and SRSF2 P95R were similar (0.37 and 0.41, respectively; supplemental Tables 3 and 4) indicating that they co-occurred in most cells of the neoplastic clone. However, the archival sample did not harbor JAK2 V617F (or did so at a level below the sensitivity of our assay) but did harbor the SRSF2 P95R at a VAF of 0.06 indicating this was the clone-founding mutation.
We and others reported that JAK2 V617F was a common driver of clonal hematopoiesis6-9 ; however, although JAK2 V617F clonal hematopoiesis is likely to be the ancestor of most MPNs, our understanding of the latency and rate of clonal expansion of JAK2 V617F–positive clones are not well understood. Our findings demonstrate that although additional driver mutations can accelerate preclinical clonal expansion of JAK2 V617F clones, the process is highly variable even between individuals whose clones harbor JAK2 V617F as the sole identifiable driver and who represent the majority of MPN patients.14
A recent study used ultrasensitive sequencing to show that clonal hematopoiesis driven by acute myeloid leukemia (AML)–associated mutations was ubiquitous among healthy adults aged 50 to 60 years.15 Interestingly, most clones identified in this study were associated with loss-of-function mutations in DNMT3A and TET2, and although most persisted over long periods (>10 years), they usually exhibited only modest expansion if any. However, in contrast to studies looking at larger numbers of people with lower assay sensitivities,6-9 this study did not identify JAK2 mutant clones in any of its 20 participants.15 It is therefore possible that JAK2 V617F, a hot spot mutation that relies on a G>T transversion that is uncommon in myeloid malignancies,16 is acquired less often but has a more pronounced effect on clonal growth than many other mutations. Nevertheless, it should be noted that the high rates of clonal growth in our study were observed in individuals that (1) were younger than the average MPN patient (as they were identified because they were registered stem cell donors) and (2) did actually go on to develop MPN and are therefore unlikely to be characteristic of the general behavior of JAK2 V617F clones. Additionally, there is a possibility that some of these clones may not have been the same from clonal hematopoiesis to MPN diagnosis, as MPN patients can sometimes harbor >1 JAK2 V617F clone.17 Nevertheless, clonal expansion rates varied dramatically even among the 12 individuals studied here. As certain germ line polymorphisms are associated with an increased risk of developing both JAK2 V617F–driven clonal hematopoiesis and MPN,11,12,17,18 we genotyped our patients for these (supplementary Table 4). In our small group of patients, we observed a possible association between the JAK2 46/1 risk haplotype (C) and the average annual rise in JAK2 V617F VAF. In fact, the 4 fastest-expanding clones were either CC homozygous in the germ line (P2 and P4) or through somatic loss of heterozygosity (P1), or had a second driver mutation (P3). These observations suggest that the 46/1 haplotype may influence the rate of expansion of established JAK2 V617F mutant clones and that loss of heterozygosity involving the risk haplotype may further expedite clonal expansion. These findings need to be verified in larger studies.
Interestingly, 1 of our patients acquired JAK2 V617F after SRSF2 p95R, and this was followed by a clonal sweep and the development of MF >12 years later. Clonal hematopoiesis driven by mutant SRSF2 is very rare before the age of 70 years,6 yet this individual harbored a significant SRSF2 P95R clone (VAF 0.06) aged only 40 years old. A recent study of 182 MF patients identified many with JAK2 V617F and SRSF2 P95 co-mutation, including several aged <70 years (n = 21, age range 39-84 years).19 Taken together, these observations suggest that younger individuals with SRSF2-mutant clonal hematopoiesis may be at a high risk of progression to a hematological neoplasm.
Our study demonstrates that JAK2 V617F neoplasms develop from clonal hematopoiesis over many years, sometimes over more than a decade. Although co-mutations in other myeloid genes may accelerate the rate of JAK2 V617F–driven clonal expansion, the rate can be highly variable even among those without co-mutations as demonstrated at least by the 7 of our 12 patients studied using targeted capture of 41 myeloid genes. Our findings suggest that heritable polymorphisms such as the JAK2 46/1 haplotype may have a role,20 but this will need to be confirmed in larger future studies and it is probable that nongenetic factors may also be at play. Our ability to predict clonal growth and by extension forecast the likelihood of progression of JAK2 V617F clonal hematopoiesis to MPN remains limited, and our study contributes to the understanding of this process. In order to better advise individuals with clonal hematopoiesis of their prognosis and to identify those that could benefit from current or future therapies, larger studies are required to define the variables influencing clonal expansion whether they are heritable vs environmental or cell-intrinsic vs cell-extrinsic and related to the hematopoietic microenvironment.21,22
The full-text version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Ayalew Tefferi for providing the numbers and age range of individuals with JAK2 V617F and SRSF2 P95 co-mutation from his team’s recent study of myelofibrosis.19
This work was supported by the Wellcome Trust Sanger Institute (WT098051). T. McKerrell is funded by a Wellcome Trust Clinician Scientist Fellowship (100678/Z/12/Z). G.S.V. is funded by a Wellcome Trust Senior Fellowship in Clinical Science (WT095663MA), and work in his laboratory is also funded by Cancer Research UK, Bloodwise, the Kay Kendall Leukaemia Fund, and Celgene. I.V. is supported by the Spanish Ministerio de Economía y Competitividad, Programa Ramón y Cajal.
Contribution: G.S.V. and P.A.C. designed the study; G.S.V., P.A.C., and T. McKerrell supervised the study, analyzed data, and wrote the manuscript with contributions from all authors; N.P., T.M., J.C., P.G., and G.C. performed experimental procedures; I.V., T. Moreno, H.P., and J.D. performed bioinformatics analyses; and K.M., C.P., M.A., and P.A.C. contributed to sample and data acquisition.
Conflict-of-interest disclosure: G.S.V. is a consultant for KYMAB and receives an educational grant from Celgene. The remaining authors declare no competing financial interests.
Correspondence: George S. Vassiliou, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, United Kingdom; e-mail: gsv20@sanger.ac.uk; and Paul A. Costeas, The Karaiskakio Foundation, Nicosia, Cyprus; e-mail: paul.costeas@karaiskakio.org.cy.