Key Points
BCR-ABL1 rearrangement as a subclonal change in ETV6-RUNX1–positive B-ALL is a rare occurrence not previously reported.
The prognosis of this rare subclonal change has not been determined, yet inclusion of tyrosine kinase inhibitors in treatment is ubiquitous.
Abstract
We report here on a case of ETV6-RUNX1–positive B-cell acute lymphoblastic leukemia (B-ALL) that has acquired a BCR-ABL1 gene rearrangement as a subclonal change. The 19-year-old female patient presented with B symptoms, pancytopenia, and circulating blasts. The bone marrow aspirate was hypercellular and was infiltrated by an immature blast population that was confirmed as B-ALL by flow cytometry. Sequential fluorescent in situ hybridization was performed on the patient’s leukemic cells, which were shown to contain both ETV6-RUNX1 and BCR-ABL1 gene rearrangements. The majority of nuclei (85%) showed only the ETV6-RUNX1 gene rearrangement; however, an additional 10% also showed a variant BCR-ABL1 gene rearrangement, indicating the ETV6-RUNX1 gene rearrangement was the primary change. A review of the literature has shown that acquisition of a BCR-ABL1 gene rearrangement as a secondary change in B-ALL is a very rare occurrence, and the effect it may have on prognosis is uncertain in the modern therapy age.
Introduction
B-cell acute lymphoblastic leukemia (B-ALL) is a neoplastic disorder of the bone marrow that represents 1.5% of all hematological malignancies.1 The average age of onset is approximately 12 years, with early peaks in childhood followed by a secondary peak in middle age. The prognostic effects of cytogenetic abnormalities in B-ALL are well known, with numerous recurring chromosome abnormalities now having been described. The most common cytogenetic abnormalities include hyperdiploidy, the cytogenetically cryptic t(12;21)(p13;q22) involving the ETV6 and RUNX1 genes, KMT2A (MLL) gene rearrangement, and the poor prognostic t(9;22)(q34;q11.2) rearrangement involving the ABL1 and BCR genes among others. The distribution of these aberrations varies markedly, depending on a patient’s age. In infant B-ALL, rearrangement of the KMT2A gene with other gene partners accounts for up to 80% of cases.2 In contrast, for adults older than 40 years, the most common cytogenetic abnormality is the t(9;22), which is seen in 20% to 30% of cases. Conversely, the t(9;22) is seen in less than 5% of the pediatric cohort.3,4 In pediatric B-ALL, the most common cytogenetic abnormality is the t(12;21), which is observed in up to 25% of all patients. The t(12;21) is rarely seen in adults.5
The t(12;21) results in fusion of the ETV6 and RUNX1 genes with production of the ETV6-RUNX1 fusion gene. The ETV6 and RUNX1 genes are both essential for normal hematopoiesis, and the production of an abnormal fusion protein may disrupt normal hematological function.4 The rearrangement may be acquired prenatally and has a variable latency period that ranges from months to years.6,7 Mouse models have shown that the presence of the ETV6-RUNX1 fusion product will not initiate leukemia; second genomic hits are required.7 The need for a second hit has been confirmed in humans, with studies showing that only 1 of 100 neonates with detectable ETV6-RUNX1 transcript at birth go on to develop ALL.7 The ETV6-RUNX1 gene rearrangement may be observed in conjunction with a hyperdiploid karyotype. However, the common cytogenetic subgroups of B-ALL are typically mutually exclusive; hence, it is rare to observe both ETV6-RUNX1 and BCR-ABL1 gene rearrangements together in a single abnormal clone.
Here we report a very rare case: To our knowledge, this is the first case confirmed by both fluorescent in situ hybridization (FISH) and molecular testing of ETV6-RUNX1–positive B-ALL with a BCR-ABL1 gene rearrangement as a subclonal change.
Methods
This is a case study and literature review. Patient consent was provided when the test was requested by the referring clinician, and there is no requirement for ethics approval for this type of report. No research was performed on the patient; clinical diagnostic cytogenetic testing was performed as per the Australian national guidelines by an accredited laboratory.
Patient presentation and treatment
The patient was a 19-year-old woman who presented with a 2-week history of abdominal pain, nausea and vomiting, weight loss, and night sweats in the context of being 12 weeks pregnant. Her initial full blood count showed pancytopenia with a hemoglobin of 111 g/L, platelets of 60 × 109/L, and neutrophils of 0.7 × 109/L, as well as circulating blasts that were variable in size with a very high nuclear to cytoplasmic ratio, agranular basophilic cytoplasm with vacuoles, round nucleus, and prominent nucleoli. A previous full blood count performed 3 weeks earlier was normal.
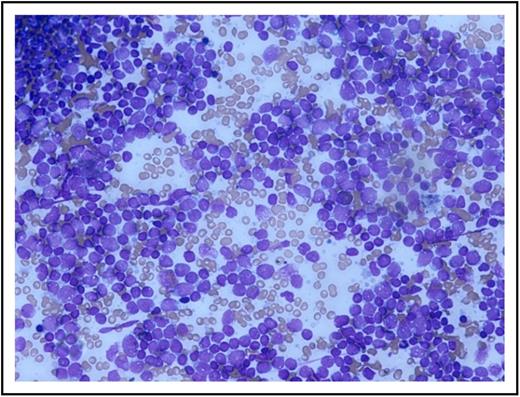
A bone marrow aspirate and trephine biopsy were performed that showed a markedly hypercellular marrow with a reduction in normal trilineage hematopoiesis with infiltration by an abnormal population of lymphoblasts (87%; Figure 1).
Patient’s bone marrow aspirate smear using May-Grünwald Giemsa stain. Image captured using Olympus microscope and camera. Original magnification ×200.
Patient’s bone marrow aspirate smear using May-Grünwald Giemsa stain. Image captured using Olympus microscope and camera. Original magnification ×200.
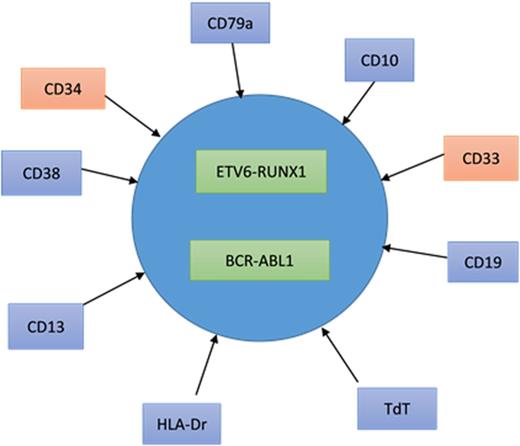
Flow cytometry on whole lysed bone marrow showed 94% blasts that revealed positivity for terminal deoxynucleotidyl transferase, human leukocyte antigen-Dr, CD10, CD19, CD79a, CD38, and CD13, with a proportion of blasts (12%) also expressing CD33 and CD34. The lymphoblasts were negative for myeloperoxidase, CD20, and CD117. This immunophenotype is consistent with B-ALL.
The patient completed induction therapy, using the Hyper-CVAD protocol (cyclophosphamide, vincristine, doxorubicin, and dexamethasone) with the addition of the tyrosine kinase inhibitor (TKI) imatinib mesylate. An allogeneic bone marrow transplant will be performed if a suitable donor is identified.
Cytogenetic and molecular cytogenetic studies
Cytogenetic studies were performed on diagnostic bone marrow, using standard techniques. Three independent cultures were established, and harvested cells were treated with trypsin and Leishman stain before analysis. A total of 20 metaphase cells were analyzed.
FISH studies were performed on fixed cultured cells, using a standard B-ALL probe panel with 200 cells scored for each probe set. The probes used initially were a KMT2A (MLL) dual-color break-apart probe, a TEL-AML1 (ETV6-RUNX1) dual-color single-fusion probe, a BCR-ABL1 dual-color dual-fusion probe, and a mixture of 3 centromeric probes for chromosomes 4, 10, and 17 to identify hyperdiploidy (Abbott Laboratories, Abbott Park, IL).
After the identification of both the t(12;21) and the t(9;22), additional FISH testing was carried out by performing sequential hybridization with an ETV6 dual-color break-apart probe (Abbott Laboratories, Abbott Park, IL) and a BCR-ABL1 dual-color dual-fusion probe (MetaSystems, Altlussheim, Germany)
Molecular studies
Total RNA was isolated from both bone marrow and peripheral blood, using automated magnetic bead extraction (Roche Magnapure). Qualitative reverse transcription polymerase chain reaction was performed to identify the BCR-ABL1 transcript, using published methods.8,9 In addition, quantitative analysis of BCR-ABL1 was performed on bone marrow, and ETV6-RUNX1 was performed on peripheral blood.8,9 Of note, the use of both bone marrow and peripheral blood for quantitative analysis was a result of a paucity of material available from bone marrow.
Results
Cytogenetics and molecular cytogenetics
Conventional cytogenetic analysis showed an abnormal hyperdiploid karyotype with structural and numerical chromosome aberrations. Abnormalities included deletion of the long arm of chromosome 6 from band q22, a deletion of the long arm of chromosome 11 from band q13 to q23, and a gain of chromosome 21 (Figure 2).
Conventional karyotype showing deletion of 6q, 11q, and trisomy of chromosome 21. The conventional karyotype according to the International Society of Cytogenetic Nomenclature 2013 was 47,XX,del(6)(q22),del(11)(q13q23),+21[10]/46,XX[10]. Image captured using Zeiss microscope and MetaSystems image analysis software.
Conventional karyotype showing deletion of 6q, 11q, and trisomy of chromosome 21. The conventional karyotype according to the International Society of Cytogenetic Nomenclature 2013 was 47,XX,del(6)(q22),del(11)(q13q23),+21[10]/46,XX[10]. Image captured using Zeiss microscope and MetaSystems image analysis software.
The initial FISH studies showed the presence of a t(12;21)(p13;q22) rearrangement in 96% of cells examined. Two clones were observed: 1 with the standard abnormal signal pattern (seen in 5% of cells) and 1 with a gain of RUNX1 (21q22) (seen in 91% of cells), which is consistent with the gain of chromosome 21 observed by conventional cytogenetics, indicating that the clone detected by conventional cytogenetics contained the t(12;21)(p13;q22). In addition, there was a loss of 1 KMT2A (11q23) signal in 93% of cells examined, which was also reflected in the conventional cytogenetics result, with all abnormal cells showing a deletion of the long arm of chromosome 11.
FISH analysis also showed the presence of a variant t(9;22)(q34;q11.2) in 10% of cells examined. The signal pattern showed a single fusion signal with 2 copies of ABL1 (9q34) and 1 copy of BCR (22q11.2). This signal pattern was above the laboratory-determined cutoff for a single-fusion signal of 6%, and indeed contained an additional ABL1 signal, indicating that this signal pattern was not as a result of simple colocalization of signals in the nucleus. There was no evidence of a t(9;22) or any variant by conventional cytogenetics; however, only 20 metaphases were able to be examined, of which only 10 showed the abnormal karyotype, with the remaining 10 showing a normal karyotype.
FISH analysis using the hyperdiploidy probe mixture showed diploidy for chromosomes 4 and 17 in all cells, with a gain of the D10Z1 probe specific for the centromere of chromosome 10 in 10% of cells. Again, this was above the laboratory cutoff for this signal pattern (5%), and there was no evidence of a trisomy 10 clone by conventional cytogenetics.
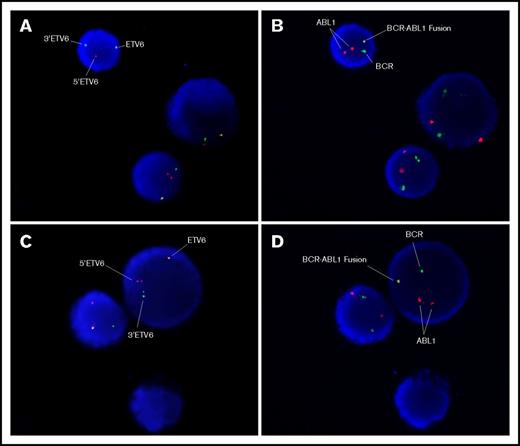
Sequential FISH studies were performed that showed that the ETV6 gene rearrangement was indeed present in cells with the BCR-ABL1 gene rearrangement, indicating that the BCR/ABL1 gene rearrangement was a subclonal change (Figure 3) and the ETV6 gene rearrangement was the primary genetic insult.
FISH images of sequential hybridization with the Vysis ETV6 break-apart probe and the MetaSystems BCR-ABL1 dual-fusion probe. The left-hand images (A,C) show the first hybridization with the ETV6 probe. The signal pattern shows 1 normal ETV6 signal and separated 3 ′ETV6 and 5 ′ETV6 signals, indicating the ETV6 gene rearrangement (t(12;21)(p13;q22)). The right-hand images (B,D) show the same cells sequentially hybridized with the BCR-ABL1 probe. The signal pattern shows 2 copies of ABL1, 1 copy of BCR, and 1 BCR-ABL1 fusion signal consistent with the BCR-ABL1 gene rearrangement, as demonstrated by molecular techniques. Image captured using Zeiss microscope and MetaSystems image analysis software.
FISH images of sequential hybridization with the Vysis ETV6 break-apart probe and the MetaSystems BCR-ABL1 dual-fusion probe. The left-hand images (A,C) show the first hybridization with the ETV6 probe. The signal pattern shows 1 normal ETV6 signal and separated 3 ′ETV6 and 5 ′ETV6 signals, indicating the ETV6 gene rearrangement (t(12;21)(p13;q22)). The right-hand images (B,D) show the same cells sequentially hybridized with the BCR-ABL1 probe. The signal pattern shows 2 copies of ABL1, 1 copy of BCR, and 1 BCR-ABL1 fusion signal consistent with the BCR-ABL1 gene rearrangement, as demonstrated by molecular techniques. Image captured using Zeiss microscope and MetaSystems image analysis software.
Molecular studies
Qualitative studies of the BCR-ABL1 transcript showed the e1a2 (p190) transcript, which is the most common form observed in B-ALL. Quantitative studies showed the diagnostic BCR-ABL/BCR ratio to be 2.7% and the ETV6-RUNX1/GUSB ratio to be 57.9%, in keeping with the relative clone abundance identified by FISH.
Discussion
The t(12;21) resulting in fusion of ETV6 and RUNX1 is the most common cytogenetic rearrangement in pediatric B-cell ALL. The ETV6-RUNX1 gene rearrangement has been shown to be in utero in origin, most likely in a B-cell progenitor, in the majority of patients; hence, its predominance in pediatric B-ALL, rather than adult B-ALL.10-12 However, the presence of ETV6-RUNX1 fusion transcripts in the Guthrie blood spots of individuals at birth greatly exceeds the number of patients who go on to develop B-ALL,13 indicating the need for additional genetic events, or second hits, for the development of clinically overt B-ALL.13 Studies have also shown that these additional genetic events can have an effect on the traditionally favorable prognosis of ETV6-RUNX1–positive B-ALL.10
There are an increasing number of common secondary abnormalities in ETV6-RUNX1–positive B-ALL that may be observed by conventional or molecular cytogenetics methods (Table 1).5,7,10,12,14-18 The vast majority of secondary abnormalities are genomic losses, with the ratio of deletion to duplication being 2:1.18
Common (>5% frequency) secondary genetic abnormalities observed in ETV6-RUNX1–positive leukemia using various methods, including conventional cytogenetics, microarray, and FISH
| Secondary abnormality . | Frequency (%) . | Possible candidate genes . |
|---|---|---|
| 12p abnormalities* | 39 | ETV6, CDKN1B |
| 9p deletion (9p13-24) including aUPD | 9-25 | CDKN2A, CDKN2B, PAX5, JAK2, MTAP |
| 12q21.33 deletion† | 13-25 | BTG1 |
| Trisomy 21‡ | 17-25 | RUNX1 |
| 5q31.3-33.3 deletion† | 23 | NR3C1, EBF1 |
| 14q32.33 deletion† | 21 | IGH |
| 3p21 deletion† | 21 | LIMD1 |
| 6q abnormalities including deletion | 13-18 | AIM1, PRDM1, FOXO3, CCNC |
| 7p14.1 deletion† | 18 | IKZF1 |
| 4q31.23 deletion† | 17 | NR3C1, ARHGAP10 |
| Trisomy 10 | 5-15 | — |
| 3q26.32 deletion† | 3-15 | TBL1XR1 |
| 3q13.2 deletion† | 15 | CD200, BTLA |
| 19q13.11 deletion† | 13 | CEBPA |
| 22q11.2 deletion† | 13 | — |
| Xq duplication (in males)† | 11 | SPANXB, HMGB3, FAM50A, HTATSF1 |
| 1q31.3 deletion† | 10 | TROVE2, GLRX2, CDC73, B3GALT2 |
| 15q15.1 abnormalities including deletion | 10 | LTK, MIRN626 |
| 13q14-34 abnormalities including deletion | 6-10 | RB1, SERP2, DLEU2, ST13P4, TRIM13, KCNRG, MIRN16-1, MIRN15A, DLEU1, DLEU7 |
| 2p25.3 deletion† | 9 | — |
| 3p14.2 deletion† | 9 | FHIT |
| X chromosome loss (in females) | 8 | — |
| 11q abnormalities including deletion | 6 | ATM, KMT2A |
| 8p11.23 deletion† | 6 | — |
| Trisomy 16 | 6 | — |
| Xp duplication† | 5 | — |
| 14q11.2 deletion† | 8 | — |
| 7q34 deletion† | 6 | NAMPT |
| Secondary abnormality . | Frequency (%) . | Possible candidate genes . |
|---|---|---|
| 12p abnormalities* | 39 | ETV6, CDKN1B |
| 9p deletion (9p13-24) including aUPD | 9-25 | CDKN2A, CDKN2B, PAX5, JAK2, MTAP |
| 12q21.33 deletion† | 13-25 | BTG1 |
| Trisomy 21‡ | 17-25 | RUNX1 |
| 5q31.3-33.3 deletion† | 23 | NR3C1, EBF1 |
| 14q32.33 deletion† | 21 | IGH |
| 3p21 deletion† | 21 | LIMD1 |
| 6q abnormalities including deletion | 13-18 | AIM1, PRDM1, FOXO3, CCNC |
| 7p14.1 deletion† | 18 | IKZF1 |
| 4q31.23 deletion† | 17 | NR3C1, ARHGAP10 |
| Trisomy 10 | 5-15 | — |
| 3q26.32 deletion† | 3-15 | TBL1XR1 |
| 3q13.2 deletion† | 15 | CD200, BTLA |
| 19q13.11 deletion† | 13 | CEBPA |
| 22q11.2 deletion† | 13 | — |
| Xq duplication (in males)† | 11 | SPANXB, HMGB3, FAM50A, HTATSF1 |
| 1q31.3 deletion† | 10 | TROVE2, GLRX2, CDC73, B3GALT2 |
| 15q15.1 abnormalities including deletion | 10 | LTK, MIRN626 |
| 13q14-34 abnormalities including deletion | 6-10 | RB1, SERP2, DLEU2, ST13P4, TRIM13, KCNRG, MIRN16-1, MIRN15A, DLEU1, DLEU7 |
| 2p25.3 deletion† | 9 | — |
| 3p14.2 deletion† | 9 | FHIT |
| X chromosome loss (in females) | 8 | — |
| 11q abnormalities including deletion | 6 | ATM, KMT2A |
| 8p11.23 deletion† | 6 | — |
| Trisomy 16 | 6 | — |
| Xp duplication† | 5 | — |
| 14q11.2 deletion† | 8 | — |
| 7q34 deletion† | 6 | NAMPT |
aUPD, acquired uniparental disomy.
12p abnormalities include deletions and translocations [not including the t(12;21)] visible, using conventional cytogenetics and deletions detected using FISH or microarray technologies.
Deletions and duplications only detected using microarray techniques, and hence not visible via conventional cytogenetics.
The additional copy of chromosome 21 may be either a normal chromosome 21 or an additional copy of the der(21)t(12;21)(p13q22).
Secondary chromosome abnormalities are detected in up to 85% of ETV6-RUNX1–positive cases. The most common findings are abnormalities of 12p (39%), with deletion of the ETV6 allele on the chromosome 12 not involved in the t(12;21) usually affected.5,7,10,12,14-17 Another common secondary change is gain of chromosome 21, seen in up to 25% of cases. Typically, the gain is of a normal chromosome 21 (70%), with a portion showing a gain of the der(21)t(12;21)(p13;q22) (30%).10 Abnormalities of 9p are seen in up to 25% of cases, and this includes cytogenetically visible deletions and submicroscopic copy number changes typically involving the tumor suppressor CDKN2A/CDKN2B and the B-cell differentiation regulator PAX5.5,10,12-17
An increasing number of submicroscopic changes have been shown to occur at relatively high frequency in ETV6-RUNX1–positive B-ALL by microarray techniques, and a number of putative causative genes have been identified. These genes are of many different classes and include genes encoding proteins involved in the cell cycle checkpoint pathway (ATM), cell cycle control (CDKN1B and CEBPA), and tumor suppression (CDKN2A, MTAP, and RB1), to name a few.5,7,12,14-18
Our patient showed a number of the common ETV6-RUNX1–positive secondary changes in the primary clone; namely, trisomy of chromosome 21 and deletion of both 6q and 11q. These abnormalities likely represent the required second genomic hit that initiated B-ALL in our patient.
In addition to the common secondary changes described earlier, a subclonal population showed the presence of concurrent BCR-ABL1 and ETV6-RUNX1 gene rearrangements in 10% of nuclei. The presence of both the BCR-ABL1 and ETV6-RUNX1 gene rearrangements indicates that the BCR-ABL1 gene rearrangement is a subclonal change in this patient’s leukemic cells that has been acquired through clonal evolution of the ETV6-RUNX1–positive leukemic clone.
The FISH pattern seen using the BCR-ABL1 probe set showed a variant signal pattern to that typically observed in a standard t(9;22)(q34;q11.2) with 1 fusion signal, 2 copies of ABL1, and 1 copy of BCR for which there are a number of possible explanations. One possibility is that the 3′ portion of the ABL1 gene has inserted adjacent to the BCR gene, resulting in a cytogenetically cryptic BCR-ABL1 gene rearrangement. Although rare, approximately 1% of all BCR-ABL1–positive chronic myeloid leukemia cases have a cytogenetically cryptic BCR-ABL1 gene rearrangement that has arisen through insertion or other, more complex, means.19 Insertional events are not restricted to BCR-ABL1, they have been reported for many genes, including KMT2A and others.20 Another possibility is that there has been a classic t(9;22)(q34;q11.2), with a deletion of BCR from 1 derivative chromosome; however, this was not observed by conventional cytogenetics as both chromosomes 9 and 22 were apparently normal. Regardless of the mechanism, molecular studies have found the BCR/ABL1 gene rearrangement to be of the e1a2 form generating the p190 protein, which is found in the majority of BCR/ABL1-positive B-ALL cases. It should also be noted that the majority of cases that have acquired the t(9;22) as a subclonal change also have the e1a2 form.21
The diagnostic level of the ETV6-RUNX1 transcript was within the typical diagnostic range, at 57.9%, whereas the BCR-ABL1 transcript was relatively low at 2.7% when compared with cases in which the primary abnormality is the t(9;22). At diagnosis, the transcript levels for BCR-ABL1–positive B-ALL can range from 35% to 138% with an average of 80%.9 The lower diagnostic level of BCR-ABL1 transcript is most likely a result of the lower percentage of leukemic cells containing the BCR-ABL1 rearrangement.
The t(9;22)(q34;q11.2) has been described as a subclonal change in a number of hematological malignancies, most predominantly in the acute myeloid leukemias; however, a number of cases of B-ALL have been reported. The acute myeloid leukemia cases reported have shown a number of recurring primary changes including inv(16)(p13q22), t(8;21)(q22;q22), and t(15;17)(q22;q12).21,22 A review of the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer showed a total of 1089 cases of t(9;22)(q34;q11.2)-positive ALL.23 On review, there were only 6 cases (0.6%) in which the t(9;22) had occurred as a secondary change (Table 2). As can be seen in Table 2, there are no consistent chromosome anomalies associated with BCR-ABL1 as a subclonal change in B-ALL. In addition to the cases listed in Table 2, there are also a small number in which a BCR-ABL1–positive clone has presented at disease relapse of ALL. In a number of these cases, however, diagnostic BCR-ABL1 testing was not able to be performed using molecular methods, and hence the true diagnostic BCR-ABL1 status is unknown.24 In B-ALL, there has been a single case report in which transcripts of both ETV6-RUNX1 and BCR-ABL1 have been detected at diagnosis. The testing was via molecular analysis, as both cytogenetic and molecular cytogenetic testing were unsuccessful, and as such, it was not possible to determine which gene rearrangement was the initiating event or, indeed, whether there were 2 de novo diseases present.25
Acute lymphoblastic leukemia cases with t(9;22)(q34;q11.2) as a subclonal change at diagnosis, taken from the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer
| Diagnostic karyotype | Publication |
| 45,XY,-7/45,idem,t(9;22)(q34;q11.2) | Uckun et al27 |
| 47,XX,+17,t(11;14)(p13;q13)/ 47,idem,t(9;22)(q34;q11.2) | Suryanarayan et al28 |
| 46,XY,t(7;11)(q36;p31)/46,idem,t(9;22)(q34;q11.2)* | Shikano et al29 |
| 45,XY,dic(9;20)(p13;q11)/45,idem, t(9;22)(q34;q11) | Safavi et al30 |
| 46,XY,t(1;7)(p13;q33)/47,idem,t(9;22)(q34;q11),+22 | Arano-Trejo et al31 |
| 46,XX,t(4;11)(q21;q23)/47,idem,+mar/49,idem,+3,+8,t(9;22)(q34;q11.2),+10,+12,+13/47-59,idem,+1,+8,t(9;22),+10,+13,+21 | Arano-Trejo et al31 |
| Diagnostic karyotype | Publication |
| 45,XY,-7/45,idem,t(9;22)(q34;q11.2) | Uckun et al27 |
| 47,XX,+17,t(11;14)(p13;q13)/ 47,idem,t(9;22)(q34;q11.2) | Suryanarayan et al28 |
| 46,XY,t(7;11)(q36;p31)/46,idem,t(9;22)(q34;q11.2)* | Shikano et al29 |
| 45,XY,dic(9;20)(p13;q11)/45,idem, t(9;22)(q34;q11) | Safavi et al30 |
| 46,XY,t(1;7)(p13;q33)/47,idem,t(9;22)(q34;q11),+22 | Arano-Trejo et al31 |
| 46,XX,t(4;11)(q21;q23)/47,idem,+mar/49,idem,+3,+8,t(9;22)(q34;q11.2),+10,+12,+13/47-59,idem,+1,+8,t(9;22),+10,+13,+21 | Arano-Trejo et al31 |
Diagnosed as T-ALL.24
The acquisition of a BCR-ABL1 gene rearrangement as a subclonal change suggests a role for BCR-ABL1 in clonal evolution and disease progression in these cases. The aberrant tyrosine kinase generated plays a role in cellular proliferation, and hence may result in a proliferative advantage to those leukemic cells containing the BCR-ABL1 rearrangement,2,22 and indeed an increased resistance to the mode of treatment often used in lower-risk B-ALL, including ETV6-RUNX1–positive cases. The possibility that the BCR-ABL1–containing cells confer a resistance to therapy may be evident in the portion of those cases that show evidence of a BCR-ABL1–positive clone detected only at relapse. It may be possible that low levels of the BCR-ABL1–positive clone were present at diagnosis, but in numbers that were below the level of detection at that time. Conventional chemotherapy, in the absence of therapeutically effective TKIs such as imatinib mesylate, may have eradicated those cells that were sensitive to chemotherapy, leaving the TKI-sensitive BCR-ABL1–positive clones to replicate and acquire new genetic changes with a potential proliferative advantage.
In B-ALL, the ETV6-RUNX1 gene rearrangement is associated with a favorable prognosis, with event-free survival approaching 90%.2,5,12,16 In contrast, the BCR-ABL1 gene rearrangement is associated with a relatively poor prognosis in both pediatric and adult cases.26 As the prognostic outcome is quite disparate when comparing ETV6-RUNX1–positive and BCR-ABL1–positive groups, there is also a difference in the intensity of treatment used for these 2 groups. The BCR-ABL1–positive patients use TKIs, with allogeneic hematopoietic stem cell transplant being recommended for suitable patients.26
The effect of a subclonal BCR-ABL1 gene rearrangement on prognosis has not been well defined because of the relative scarcity of cases across all subtypes of acute leukemia. However, in the few acute myeloid leukemia cases reported with a BCR-ABL1 gene rearrangement as a subclonal change, a number with the prognostically favorable inv(16) have shown outcomes in line with that expected of a typical inv(16), suggesting that the BCR-ABL1 gene rearrangement may have no effect on prognosis in these cases. It should, however, be noted that many of these patients have been treated with TKIs as a part of the therapeutic regimen.
The 6 cases of ALL with BCR/ABL1 as a secondary change have very limited or no available data on both treatment type and event-free survival. In the single case reported that showed both ETV6-RUNX1 and BCR-ABL1 transcripts, the therapeutic regimen used was according to the AIEOP-BFM-ALL2000 protocol; namely, methylprednisolone, vincristine, farmorubicin, and l-asparaginase in combination with the TKI imatinib mesylate. Remission was achieved after the first induction with residual disease of 0.035% for ETV6/RUNX1 and 0.023% for BCR/ABL1 after 12 months with no adverse events reported.25
Using the Hyper-CVAD protocol with imatinib mesylate, our patient has achieved a morphologic and cytogenetic remission after the first induction cycle with residual undetectable BCR-ABL1/ABL and ETV6-RUNX1/GUSB of 0.16%.
In conclusion, to the best of our knowledge, this patient is the first proven case of BCR-ABL1 gene rearrangement acquired as a subclonal change in ETV6-RUNX1–positive leukemia. The prognosis for these rare patients with subclonal BCR-ABL1 gene rearrangements is as yet unclear, particularly when treated with a modern treatment regime that includes the addition of TKI therapy, and more cases will need to be documented to determine the most appropriate treatment regimen and likely prognostic implications for this very small subgroup of B-ALL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
Quantitative ETV6-RUNX1 assays were performed by Elizabeth Algar (Genetics and Molecular Pathology, Monash Health, Victoria, Australia).
Authorship
Contribution: K.A.D. wrote the manuscript; K.A.D., R.V., T.J.B., L.A.R., G.D., and J.W. performed research; all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karen A. Dun, Cytogenetics Laboratory, Pathology Services, Royal Hobart Hospital, 48 Liverpool St, Hobart, TAS 7000, Australia. e-mail: karen.dun@ths.tas.gov.au.

![Figure 2. Conventional karyotype showing deletion of 6q, 11q, and trisomy of chromosome 21. The conventional karyotype according to the International Society of Cytogenetic Nomenclature 2013 was 47,XX,del(6)(q22),del(11)(q13q23),+21[10]/46,XX[10]. Image captured using Zeiss microscope and MetaSystems image analysis software.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/2/10.1182_bloodadvances.2016000463/3/m_advances000463f2.jpeg?Expires=1771328571&Signature=mKtIVb~cAJTtHkWXmxcJ0c8E8UQ5wYw3QJaNuLuWl59KBBuHmM8-02RfBnG8KkZjZXnTiRf7JXSvmuaDXZjWvAJUCcZ-Oyj7ve8niDXhLPnbOoIOoFTRq6LNrZGeltrMzhvSZyks3lSbNJ~Y1iIrXS15C~T2lcInu23l79hXsu33r~MH7CLQAhvnzneSYrITFzuTH7v~mgck-UZ6ChEE4pxkNjnHQYgHcgfUrtCygHqIXaW~og5mOsp5jWuXQD3pXM9bduE3kSE6K3z8naxzLqLEB4a8Mgy5E2FAjQNvXUFZH0avTCGhRZj1ZXKajn0LML5on8lbXqZ2uHln3D8fug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)