Key Points
The majority of kinase alterations identified in Ph-like ALL can be targeted with either ABL or JAK inhibition.
Enhanced efficacy of tyrosine kinase inhibitor therapy was observed with dexamethasone in patient-derived xenograft models of Ph-like ALL.
Abstract
New therapies for Philadelphia chromosome–like acute lymphoblastic leukemia (Ph-like ALL) patients are urgently needed. The genetic landscape of Ph-like ALL is characterized by a diverse array of kinase-activating alterations (including rearrangements, sequence mutations, and copy number alterations), suggesting that patients with Ph-like ALL are candidates for targeted therapy, similar to BCR-ABL1 ALL. We sought to investigate the functional role and targetability of the spectrum of kinase-activating alterations identified in Ph-like ALL. We demonstrate cytokine-independent growth and activation of JAK-STAT signaling pathways in Ba/F3 cells by all alterations tested. The development of murine Arf−/− pre-B ALL expressing RCSD1-ABL2 or SSBP2-CSF1R was accelerated with the presence of IK6, a dominant negative isoform of Ikaros common in Ph-like ALL, providing evidence that these fusions are leukemogenic. In vitro screening using a panel of tyrosine kinase inhibitors against 14 different kinase alterations identified the ABL1-inhibitor, dasatinib, as a potent inhibitor of ABL-class fusions (ABL1, ABL2, CSF1R, PDGFRB), whereas the JAK1/JAK2 inhibitor ruxolitinib, was most effective against JAK-STAT–activating alterations (JAK1, JAK2, JAK3, IL7R, IL2RB), but not TYK2. Evaluation of dasatinib or ruxolitinib against patient-derived xenograft models demonstrated superior antileukemic efficacy when combined with dexamethasone compared with either agent alone. These data provide the foundation for rationally designed clinical trials that assess the efficacy of targeted therapy in patients with Ph-like ALL.
Introduction
Philadelphia chromosome–like acute lymphoblastic leukemia (Ph-like ALL) or BCR-ABL1–like is a high-risk subtype of childhood and adult ALL. Ph-like ALL patients are defined as having a gene expression signature similar to BCR-ABL1–positive ALL, but lack the BCR-ABL1 fusion gene, and commonly harbor genetic alterations targeting B-lymphoid transcription factors, including IKZF1 (Ikaros).1,2 The prevalence of Ph-like ALL rises from 10% in standard-risk childhood ALL to more than 25% in young adults (21-39 years) and adults (>40 years), and this subtype is associated with a poor outcome in both children and adults.3-6
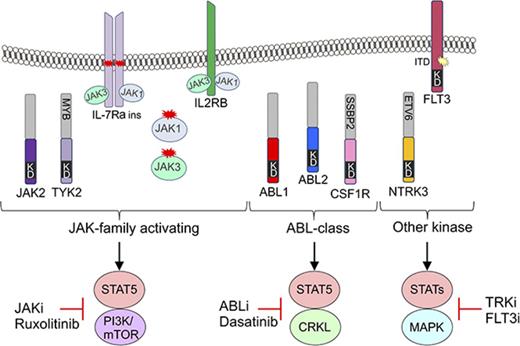
Fusion genes involving 17 cytokine receptor or tyrosine kinases have been identified in patients with Ph-like ALL.3,4,7,8 Approximately 50% of patients harbor rearrangements activating cytokine receptor like factor 2 (CRLF2), with frequent concomitant sequence mutations in Janus kinases or other regulators of JAK-STAT signaling, particularly JAK2,9-12 that are potentially amenable to treatment with JAK inhibitors (JAKis) such as ruxolitinib.13,14 Approximately one-third of Ph-like non-CRLF2 ALL patients harbor chromosomal rearrangements that result in either deregulation of a cytokine receptor or the formation of kinase fusion genes.3,4,7,8 A major subgroup includes those that are predicted to respond to ABL1 inhibitors (ABLis), such as imatinib and dasatinib, with rearrangements involving ABL1 (n = 12 fusion partners), ABL2 (n = 3), CSF1R (n = 3), LYN (n = 2), PDGFRA (n = 1), and PDFGRB (n = 7). A second major group includes rearrangements that activate JAK family kinases, including JAK2 (n = 20), EPOR (n = 4), and TYK2 and IL2RB (n = 1 each). A third group constitutes a variety of other kinases or cytokine receptors, including NTRK3, FLT3, FGFR1, and BLNK, all with 1 fusion partner thus far identified. In addition to kinase-activating alterations, Ph-like ALL patients harbor loss-of-function alterations in the tumor suppressors IKZF1 (∼70%) and CDKN2A/B (encoding Arf; ∼50%), which may also influence treatment response to conventional chemotherapy.15,16
We have previously demonstrated in vitro activity of dasatinib against a limited number of ABL1-class fusions expressed in Arf−/− pre-B cells (EBF1-PDGFRB, RCSD1-ABL2, and SSBP2-CSF1R).3,8 However, the role of these alterations in leukemia development and sensitivity to tyrosine kinase inhibitors (TKIs) in vivo has not been assessed. Furthermore, variable efficacy of the JAK inhibitor ruxolitinib has been observed for human leukemic cells harboring CRLF2 or EPOR rearrangements,13,17 and the targetability of novel rearrangements involving TYK2 and IL2RB is unknown. We sought to investigate the functional role of kinase fusions identified in Ph-like ALL and screen a panel of TKIs against a diverse selection of Ph-like ALL alterations to identify the most potent and specific agent for each alteration that can be tested in clinical trials designed to improve the outcome of Ph-like ALL.
Methods
Gene expression profiling
For mouse cells, whole bone marrow was harvested from wild-type C57Bl/6 mice and flow sorted into Hardy fractions of B-cell development based on surface marker expression (supplemental Table 2). Gene expression profiling was performed using MG-430 2.0 microarrays according to the manufacturer’s instructions (Affymetrix). Signals were scaled to a median value of 500 using the Affymetrix MAS 5.0 algorithm. Probe sets with absent calls were excluded, and probe set signals were variance-stabilized by adding the number 32 and subsequently log2 transformed. Gene expression data for normal human hematopoiesis (HemaExplorer) were downloaded as log2 transformed from BloodSpot.18 Unsupervised hierarchical clustering was performed using R package.
Retroviral constructs, cell culture, and TKI screening
Subcloning of full-length fusions into MSCV-ires-GFP, retroviral infection, cell culture, and screening of TKI compounds were performed as previously described.8 Details are provided in the supplemental Appendix.
Phosphoflow analysis
To assess phosphorylation, cells were adjusted to 1 × 106 cells/mL and treated with or without inhibitors for 1 hour. Cells were subsequently fixed, permeabilized, and stained with either anti-STAT5 (pY694)-Ax647 (BD Biosciences) or anti-CRKL (pY207; Cell Signaling Technology) followed by anti-rabbit Pacific Blue conjugated anti-mouse immunoglobulin G (IgG) secondary antibody (Life Technologies). Patient-derived xenograft (PDX) cells were also stained with antisera to human CD45-BV605 and human CD19-PE (BD Biosciences) to gate on leukemic blasts. Cells were collected on an LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo (Tree Star) and Cytobank (www.cytobank.org).
Mouse pre–B-cell transplantation assays
Arf−/− pre-B cells were retrovirally transduced with MSCV-ires-GFP, MSCV-p185 BCR-ABL1-ires-GFP, MSCV-RCSD1-ABL2-ires-GFP, or MSCV-SSBP2-CSF1R-ires-GFP with MSCV-IK6-ires-RFP. For transplantation experiments, 5 × 105 GFP/RFP double-positive sorted cells were IV injected into sublethally irradiated (5 Gy) C57Bl/6 recipient mice. Animals were monitored daily for clinical signs of leukemia and euthanized when moribund. Immunophenotyping was performed on bone marrow for B220, CD19, CD43, BP1, IgM, CD3, Mac-1, Gr-1, Ter119, and Thy1 (BD Biosciences). All experiments were performed on protocols approved by the St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee.
Patient-derived xenograft models
Xenograft models of human Ph-like ALL were established using diagnostic samples (bone marrow or peripheral blood) from patients treated on St. Jude Children’s Research Hospital or the Children’s Oncology Group protocols. Briefly, primary leukemia cells (106 cells per mouse) were transplanted via tail vein injection into sublethally irradiated (2.5 Gy) nonobese diabetic (NOD).Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NOD-severe combined immunodeficiency γ-null [NSG]) mice. Bone marrow harvested from engrafted mice was transduced with lentivirus expressing YFP and luciferase to enable bioluminescent imaging using a Xenogen IVIS-200 system and Living Image software (Caliper Life Sciences), as previously described.19 Details for the in vivo treatment studies are provided in the supplemental Appendix. All experiments were performed on protocols approved by the St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee.
Results
Kinase fusions induce cytokine-independent growth through pSTAT5 and pCRKL
Collectively, more than 60 different kinase-activating rearrangements have been identified in patients with Ph-like B-ALL (supplemental Table 1).3,4 Because the expression of the kinases altered in Ph-like ALL is tightly regulated during normal hematopoiesis, we hypothesized that their overexpression and constitutive activation in lymphoid leukemic blasts was due to fusion with 5′ partner genes that are expressed during B-cell differentiation and facilitate constitutive kinase activation. To investigate this, we analyzed gene expression profiling on populations of mouse and human hematopoietic cells that recapitulate normal ontogeny. All 33 of the 5′ genes investigated in this study were moderately to highly expressed during mouse B-cell development. In contrast, the expression of all 9 kinase and cytokine receptor genes was low in normal B cells obtained from mouse and human bone marrow, indicating that fusion to 5′ fusion partners or juxtaposition to strong enhancers results in deregulation in the B-cell lineage (Figure 1).
Expression of 5′ and 3′ fusion partner genes during normal ontogeny. Unsupervised clustering of gene expression data from (A) mouse and (B) human hematopoietic cells. Kinase genes are highlighted in red. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; DC, dendritic cell; GMP, granulocyte monocyte progenitor; HPC, hematopoietic progenitor cell; HSC, hematopoietic stem cell; MEP, megakaryocyte-erythroid progenitor; mono, CD14+ monocyte; NK, natural killer.
Expression of 5′ and 3′ fusion partner genes during normal ontogeny. Unsupervised clustering of gene expression data from (A) mouse and (B) human hematopoietic cells. Kinase genes are highlighted in red. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; DC, dendritic cell; GMP, granulocyte monocyte progenitor; HPC, hematopoietic progenitor cell; HSC, hematopoietic stem cell; MEP, megakaryocyte-erythroid progenitor; mono, CD14+ monocyte; NK, natural killer.
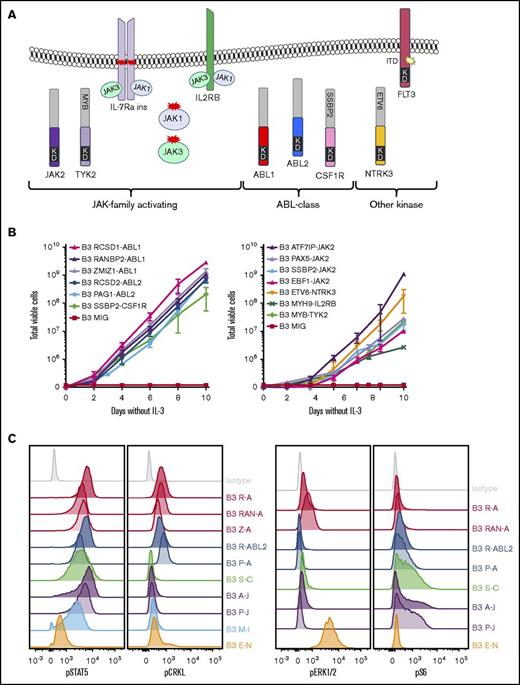
To investigate the functional role of these alterations in Ph-like ALL, we expressed 13 kinase fusions involving either ABL1-class, JAK-family, or other kinase genes into mouse interleukin-3 (IL-3)–dependent Ba/F3 or IL-7–dependent Arf−/− pre-B cells.20 The specific fusions studied were ABL1 (RCSD1-ABL1, RANBP2-ABL1, ZMIZ1-ABL1), ABL2 (RCSD1-ABL2, PAG1-ABL2), CSF1R (SSBP2-CSF1R), JAK2 (ATF7IP-JAK2, EBF1-JAK2, PAX5-JAK2, SSBP2-JAK2), TYK2 (MYB-TYK2), IL2RB (MYH9-IL2RB), and NTRK3 (ETV6-NTRK3) (Figure 2A). Expression of each fusion was confirmed by quantitative reverse transcription polymerase chain reaction (RT-PCR) (supplemental Figure 1; supplemental Table 3). All fusions tested were in-frame and retained the intact tyrosine kinase domain of the 3′ fusion partner and conferred the ability of Ba/F3 cells to survive and proliferate in the absence of IL-3 (Figure 2B). Empty vector transduced cells did not exhibit cytokine-independent proliferation.
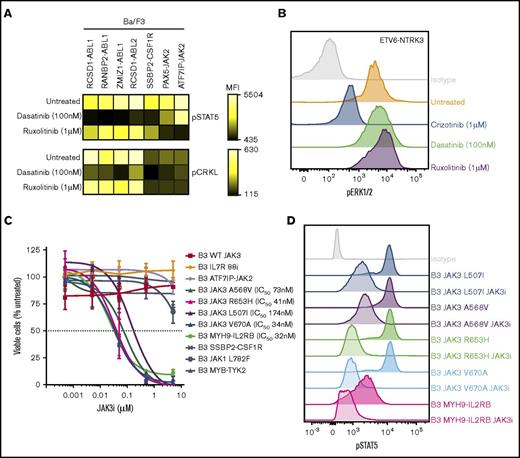
Cytokine independence and constitutive activation of signaling pathways in Ba/F3 cells. (A) Kinase alterations investigated in this study. Red and yellow shapes denote sequence mutations. (B) Growth of Ba/F3 cells in the absence of IL-3. Cell number was determined using trypan blue. Each point represents the mean ± standard deviation (SD; n = 3). (C) Phosphoflow cytometric analysis of signaling phosphoproteins in Ba/F3 cells. Cells were grown in the absence of IL-3, harvested, and assessed for the phosphorylation of pSTAT5, pCRKL, pERK1/2, and PS6. A-J, ATF7IP-JAK2; E-N, ETV6-NTRK3; KD, kinase domain; M-I, MYH9-IL2RB; P-A, PAG1-ABL2; P-J, PAX5-JAK2; R-A, RCSD1-ABL1; R-ABL2, RCSD1-ABL2; RAN-A, RANBP2-ABL1; S-C, SSBP2-CSF1R; Z-A, ZMIZ1-ABL1.
Cytokine independence and constitutive activation of signaling pathways in Ba/F3 cells. (A) Kinase alterations investigated in this study. Red and yellow shapes denote sequence mutations. (B) Growth of Ba/F3 cells in the absence of IL-3. Cell number was determined using trypan blue. Each point represents the mean ± standard deviation (SD; n = 3). (C) Phosphoflow cytometric analysis of signaling phosphoproteins in Ba/F3 cells. Cells were grown in the absence of IL-3, harvested, and assessed for the phosphorylation of pSTAT5, pCRKL, pERK1/2, and PS6. A-J, ATF7IP-JAK2; E-N, ETV6-NTRK3; KD, kinase domain; M-I, MYH9-IL2RB; P-A, PAG1-ABL2; P-J, PAX5-JAK2; R-A, RCSD1-ABL1; R-ABL2, RCSD1-ABL2; RAN-A, RANBP2-ABL1; S-C, SSBP2-CSF1R; Z-A, ZMIZ1-ABL1.
To assess activation of kinase signaling pathways, we performed phosphoflow cytometry analysis of transduced Ba/F3 cells (Figure 2C). Phosphorylation of STAT5 was detected in all cell lines, with weaker phosphorylation observed in Ba/F3 cells expressing ETV6-NTRK3. Activation of CRKL was detected only in cells expressing ABL1 or ABL2 fusions. Phosphorylation of extracellular signal-regulated kinase 1/2 was only detected in the Ba/F3-ETV6-NTRK3 cell line, confirming previous reports of MAPK pathway activation by this fusion.21 Phosphorylation of ribosomal protein S6, a downstream target of the phosphatidylinositol-3-kinase (PI3K) pathway, was variable. These data indicate that kinase fusions activate distinct but overlapping signaling pathways to induce cytokine-independent growth of Ba/F3 cells.
ABL2 and CSF1R fusions induce B-ALL in a de novo mouse model
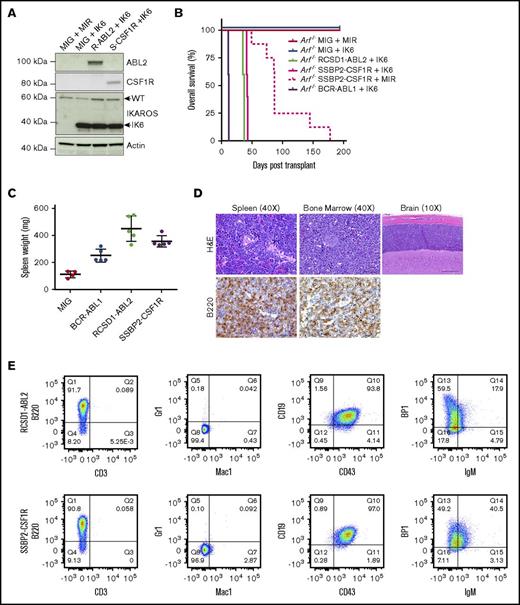
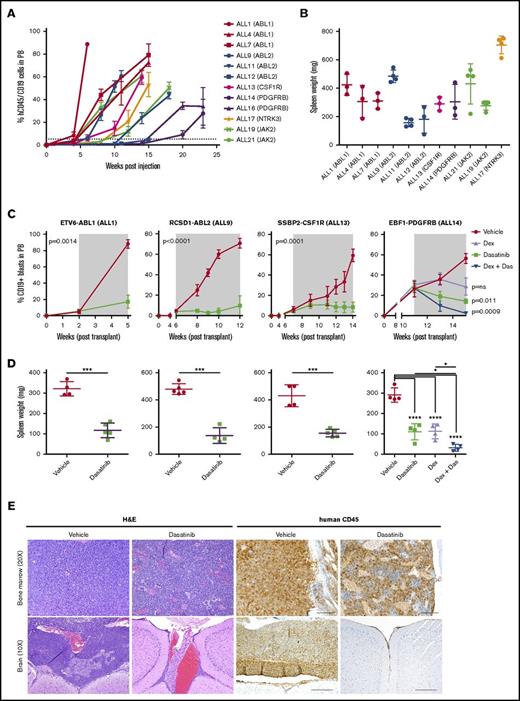
To determine whether novel kinase fusions contribute to leukemogenesis in vivo, we introduced MSCV-ires-GFP retroviral supernatants containing empty vector, p185 BCR-ABL1, RCSD1-ABL2, or SSBP2-CSF1R into IL-7–dependent Arf−/− mouse pre-B cells expressing MSCV-ires-RFP or the dominant negative isoform of Ikaros, IK6 (MSCV-ires-IK6-RFP) (Figure 3A). We chose the Arf−/− pre–B-cell system because these are primary cells at the appropriate developmental stage; deletions of CDKN2A/B are frequent in Ph-like ALL (∼50%); it provides the opportunity to comodel additional alterations; and it is an excellent in vivo model for studying the leukemogenic properties of kinase alterations identified in B-ALL, as demonstrated with BCR-ABL1 B-ALL.15,20 Mice transplanted with cells expressing RCSD1-ABL2 or SSBP2-CSF1R and IK6 developed leukemia with a median latency of 36 and 40 days, respectively, whereas pre-B cells expressing SSBP2-CSF1R alone succumbed to disease with a median latency of 87 days (Figure 3B). Mice transplanted with cells expressing empty vector were still alive up to day 300.
Expression of RCSD1-ABL2 and SSBP2-CSF1R in Arf−/−pre-B cells induces B-ALL in C57Bl/6 mice. (A) Immunoblot of Arf−/− pre-B cells showing expression of ABL2, CSF1R, IKAROS, and actin. (B) Kaplan-Meier curve of sublethally irradiated C576Bl/6 mice receiving Arf−/− pre-B cells by tail vein injection (5 × 105 cells/mouse). (C) Spleen weight of C576Bl/6 mice transplanted with Arf−/− pre-B cells (n = 5). MIG, MSCV-ires-GFP. (D) Bone marrow, spleen, and brain sections from a representative mouse transplanted with Arf−/− pre-B cells stained with hematoxylin and eosin (H&E) or a B220-specific antibody. (E) Analysis of bone marrow of diseased animals by flow cytometry. Immunophenotype is consistent with arrest at the pre-B stage of development.
Expression of RCSD1-ABL2 and SSBP2-CSF1R in Arf−/−pre-B cells induces B-ALL in C57Bl/6 mice. (A) Immunoblot of Arf−/− pre-B cells showing expression of ABL2, CSF1R, IKAROS, and actin. (B) Kaplan-Meier curve of sublethally irradiated C576Bl/6 mice receiving Arf−/− pre-B cells by tail vein injection (5 × 105 cells/mouse). (C) Spleen weight of C576Bl/6 mice transplanted with Arf−/− pre-B cells (n = 5). MIG, MSCV-ires-GFP. (D) Bone marrow, spleen, and brain sections from a representative mouse transplanted with Arf−/− pre-B cells stained with hematoxylin and eosin (H&E) or a B220-specific antibody. (E) Analysis of bone marrow of diseased animals by flow cytometry. Immunophenotype is consistent with arrest at the pre-B stage of development.
At end-stage disease, we observed a significant increase in spleen weight for tumors harboring RCSD1-ABL2 (450 ± 42 mg, n = 5; P = .0002) and SSBP2-CSF1R (355 ± 19 mg, n = 5; P < .0001) compared with empty vector controls (112 ± 12 mg, n = 5) (Figure 3C). All mice showed >70% replacement of GFP/RFP+ cells in the bone marrow with infiltration of B220+ leukemic blasts in the spleen, bone marrow, liver, and brain (Figure 3D), and a pre-B immunophenotype (CD43+, B220+, CD19+, BP-1+, and IgM−) (Figure 3E; supplemental Table 4). Thus, fusions involving constitutively activated kinases not usually expressed in B-cell progenitors, ABL2 and CSF1R, can induce the development of ALL when aberrantly expressed in pre-B cells.
Screening for TKIs that target kinase alterations in Ph-like ALL
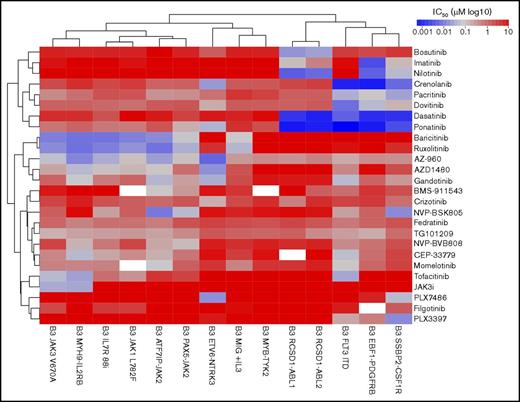
To gain a detailed understanding of the TKI sensitivity profile in Ph-like ALL, we performed a screen of 14 Ba/F3 cell lines with 26 compounds previously shown to be active against ABL1, JAK1/2/3, or type III receptor tyrosine kinases (RTKs; CSF1R, FLT3, PDFGRB). The kinase alterations tested included the ABL fusions RCSD1-ABL1 and RSCD1-ABL2; type III RTKs SSBP2-CSF1R, EBF1-PDGFRB, and FLT3 internal tandem duplication (ITD),22 the JAK-family fusions ATF7IP-JAK2, PAX5-JAK2, and MYB-TYK2; additional JAK-STAT activating alterations IL7R p.IsoLeu241-242ThrCys,23 JAK1 p.Leu782Phe,24 JAK3 p.Val670Ala,24 and MYH9-IL2RB; and ETV6-NTRK3. Cell viability was measured after 48 hours of exposure to drug, and the results are presented as 50% inhibitory concentration (IC50) values for each drug and cell line (supplemental Table 5).
Unsupervised clustering of the IC50 values grouped the cell lines based on the underlying kinase alteration (Figure 4). Specific inhibition of ABL-class fusions was observed with the ABLi imatinib (IC50 values of 33-400 nM) and nilotinib (IC50 values of 10-18 nM), whereas other cell lines were not affected up to 10 μM. The dual class SRC/ABL inhibitor dasatinib25 and the third-generation ABLi ponatinib26 were 10- to 100-fold more potent against ABL-class alterations, with IC50 values of 0.8 to 2 nM. The ABL1-specific inhibitor, bosutinib,27 was effective only against Ba/F3 cells expressing RCSD1-ABL1 or RCSD1-ABL2 fusions, with IC50 values of 0.8 to 2 nM. The efficacy of dasatinib was confirmed on an extended panel of cell lines, including RANBP2-ABL1, ZMIZ1-ABL1, and PAG1-ABL2 (supplemental Figure 2A).
In vitro sensitivity of Ba/F3 cells to tyrosine kinase inhibitors. Response of 14 Ba/F3 cell lines expressing Ph-like ALL alterations to targeted agents. Cell viability was measured after 48 hours using CellTitre-Blue viability assay. IC50 values were calculated using serial dilutions and linear regression. The heat map shows IC50 values (log10 scale) according to the color key. Cell lines (columns) and drugs (rows) were ordered by unsupervised clustering.
In vitro sensitivity of Ba/F3 cells to tyrosine kinase inhibitors. Response of 14 Ba/F3 cell lines expressing Ph-like ALL alterations to targeted agents. Cell viability was measured after 48 hours using CellTitre-Blue viability assay. IC50 values were calculated using serial dilutions and linear regression. The heat map shows IC50 values (log10 scale) according to the color key. Cell lines (columns) and drugs (rows) were ordered by unsupervised clustering.
The multikinase inhibitors crenolanib (known to target FLT3)22 and dovitinib (known to target FGFR128 and PDGFRs)29 showed efficacy against all type III RTKs tested, with IC50 values of 0.9 to 7 nM for crenolanib and 8 to 126 nM for dovitinib. We also confirmed the specificity of the CSF1R inhibitor, PLX3397, for the CSF1R fusion (IC50 value, 15.5 nM), and to a lesser extent FLT3 and PDGFRB alterations (IC50 values of 174 nM and 460 nM, respectively). Moderate efficacy of crizotinib, an ALK inhibitor,30 against ETV6-NTRK3 was confirmed (IC50 value, 143 nM)31 ; however, we identified a more potent NTRK3 inhibitor, PLX7486 (IC50 value, 17.5 nM).32
The JAK inhibitor ruxolitinib was effective against all cell lines harboring JAK-family fusions or mutations, including the MYH9-IL2RB rearrangement, with IC50 values ranging from 8 to 46 nM. These results were confirmed on an extended panel of JAK2 fusions and sequence mutations (supplemental Figure 2B). Similar sensitivities were observed for the JAK1/JAK2 inhibitor, baricitinib. The most specific type 1 JAK2 inhibitors identified in this panel were NVP-BSK80533 and NVP-BVB808,34 with low IC50 values for cells harboring JAK2 fusions (10-170 nM), moderate IC50 values for cells expressing the IL7R p.IsoLeu241-242ThrCys mutation or FLT3 ITD (both ∼200 nM), with little effect on the remaining cell lines. NVP-BSK805 was more potent compared with NVP-BVB808. The JAK3 inhibitor tofacitinib35 was specific for cells expressing the JAK3 p.Val670Ala mutation (IC50 value, 47 nM) and the MYH9-IL2RB rearrangement (IC50 value, 26 nM), but also inhibited other JAK family alterations (ATF7IP-JAK2, JAK1 p.Leu782Phe, and IL7R p.IsoLeu241-242ThrCys) at moderate concentrations (IC50 values, 63-282 nM). One striking observation was the exquisite sensitivity of cells expressing the JAK3 p.Val670Ala mutation (IC50 value, 34 nM) and MYH9-IL2RB rearrangement (IC50 value, 32 nM) to a novel JAK3 inhibitor, JAK3i,36 with no effect on other cell lines up to 10 μM. This was confirmed on an extended panel of JAK3 mutations: p.Leu507Ile, p.Ala568Val, and p.Arg653His (Figure 5C). We did not identify any compounds effective against the MYB-TYK2 fusion.
Inhibition of signaling pathways by tyrosine kinase inhibitors in Ba/F3 cells. (A) Ba/F3 cells were treated with dasatinib or ruxolitinib for 1 hour, harvested, and assessed for phosphorylation of STAT5 and CRKL. Heat map indicates mean fluorescent intensity and was generated using Cytobank. (B) Phosphorylation of extracellular signal-regulated kinase 1/2 in Ba/F3-ETV6-NTRK3 cells at basal levels or after treatment with crizotinib, dasatinib, or ruxolitinib (1 hour). (C) Dose-response curve of Ba/F3 cells after 48 hours’ treatment with JAK3i. Values are normalized to dimethyl sulfoxide controls for each cell line and represent mean ± SD (n = 3). (D) Phosphorylation of STAT5 in Ba/F3 cells at basal levels or after JAK3i treatment (1 hour).
Inhibition of signaling pathways by tyrosine kinase inhibitors in Ba/F3 cells. (A) Ba/F3 cells were treated with dasatinib or ruxolitinib for 1 hour, harvested, and assessed for phosphorylation of STAT5 and CRKL. Heat map indicates mean fluorescent intensity and was generated using Cytobank. (B) Phosphorylation of extracellular signal-regulated kinase 1/2 in Ba/F3-ETV6-NTRK3 cells at basal levels or after treatment with crizotinib, dasatinib, or ruxolitinib (1 hour). (C) Dose-response curve of Ba/F3 cells after 48 hours’ treatment with JAK3i. Values are normalized to dimethyl sulfoxide controls for each cell line and represent mean ± SD (n = 3). (D) Phosphorylation of STAT5 in Ba/F3 cells at basal levels or after JAK3i treatment (1 hour).
Inhibition of kinase signaling in Ph-like ALL
We next examined the effect of kinase inhibition on downstream signaling in Ba/F3 cells harboring kinase fusions. Treatment with dasatinib (100 nM) reduced constitutive phosphorylation of STAT5 to basal levels in Ba/F3 cells harboring ABL-class fusions, whereas ruxolitinib (1 μM) reduced pSTAT5 expression in Ba/F3 cells expressing JAK2 fusions (Figure 5A), indicating that activation of STAT5 is an important oncogenic signaling pathway for these kinase fusions. Inhibition of pCRKL by dasatinib was only observed in cells expressing ABL1 or ABL2 kinase fusions (Figure 5A). Specific inhibition of pERK1/2 was observed in Ba/F3-ETV6-NTRK3 cells treated with crizotinib (1 μM), whereas dasatinib and ruxolitinib had no affect (Figure 5B). We also observed a marked reduction in phosphorylation of STAT5 in cells expressing JAK3 mutants or the MYH9-IL2RB rearrangement treated with JAK3i (1 μM) (Figure 5D).
Establishment of patient-derived xenograft models of Ph-like ALL
The cell line studies described demonstrate in vitro activity, but do not provide evidence of efficacy on human cells in vivo. To directly examine this we established PDX models of Ph-like ALL by IV injecting 1 × 106 primary human leukemic cells carrying defined kinase lesions into NSG mice. Engraftment was assessed by measurement of human CD45+/CD19+ leukemic blasts in peripheral blood. Overall, 42 of 61 transplanted tumors engrafted, representing a variety of kinase alterations identified in Ph-like ALL (supplemental Table 6). These will serve as an important resource for the study of this subtype and are available upon request from the authors. The kinetics of engraftment differed between tumors, with the time taken to reach 5% human CD45+/19+ cells in the peripheral blood ranging from 2 to 17 weeks (Figure 6A). We also observed variation in both spleen weights (range, 135-750 mg) (Figure 6B) and central nervous system involvement for each tumor (supplemental Table 6). RT-PCR confirmed that each PDX expressed the kinase alteration present in the primary patient samples (data not shown). Primary leukemic cells harboring ABL-class fusions were sensitive to dasatinib; cells harboring JAK2 or EPOR-rearrangements were sensitive to ruxolitinib treatment ex vivo (supplemental Figure 3).
Dasatinib is effective in vivo against ABL-class fusions. (A) Levels of human CD45/CD19+ blasts in the peripheral blood of NSG mice injected with primary B-ALL cells. (B) Spleen weight of NSG mice at harvest. (C) NSG mice were injected with primary leukemic cells. Upon peripheral engraftment of 5% human CD45, animals (n = 5) were treated with vehicle, dasatinib (20 mg/kg per day), dexamethasone (4 mg/mL ad libitum), or dasatinib and dexamethasone combined. Treatment length is indicated by the gray shaded area; animals were euthanized at the last time point. Analysis of slope was measured by linear regression and unpaired 2-way t test comparing vehicle with dasatinib (ALL1, ALL9, ALL13), or 1-way analysis of variance (ANOVA) with Tukey posttest for multiple comparisons (ALL14). (D) Spleen weights were measured at the time of euthanasia. Each point represents the mean ± SD (n = 5). (E) Representative H&E and human CD45 staining of bone marrow and brain harvested from vehicle or dasatinib-treated mice of ALL1. *P < .05, ***P < .001, ****P < .0001. Das, dasatinib; dex, dexamethasone.
Dasatinib is effective in vivo against ABL-class fusions. (A) Levels of human CD45/CD19+ blasts in the peripheral blood of NSG mice injected with primary B-ALL cells. (B) Spleen weight of NSG mice at harvest. (C) NSG mice were injected with primary leukemic cells. Upon peripheral engraftment of 5% human CD45, animals (n = 5) were treated with vehicle, dasatinib (20 mg/kg per day), dexamethasone (4 mg/mL ad libitum), or dasatinib and dexamethasone combined. Treatment length is indicated by the gray shaded area; animals were euthanized at the last time point. Analysis of slope was measured by linear regression and unpaired 2-way t test comparing vehicle with dasatinib (ALL1, ALL9, ALL13), or 1-way analysis of variance (ANOVA) with Tukey posttest for multiple comparisons (ALL14). (D) Spleen weights were measured at the time of euthanasia. Each point represents the mean ± SD (n = 5). (E) Representative H&E and human CD45 staining of bone marrow and brain harvested from vehicle or dasatinib-treated mice of ALL1. *P < .05, ***P < .001, ****P < .0001. Das, dasatinib; dex, dexamethasone.
Enhanced efficacy of TKIs with dexamethasone in Ph-like ALL
To determine the efficacy of dasatinib in Ph-like ALL, we performed preclinical studies using PDX models of ETV6-ABL1, RCSD1-ABL2, SSBP2-CSF1R, or EBF1-PDGFRB B-ALL (ALL1, ALL9, ALL13, and ALL14, respectively). Mice were randomized and vehicle or dasatinib (20 mg/kg per day by mouth) treatment commenced when disease burden reached 5% human CD45+/CD19+ blasts in the peripheral blood, with a treatment duration of 5 to 6 weeks. Dasatinib-treated mice had significantly reduced leukemic burden compared with vehicle-treated mice, as assessed by lower peripheral blasts, spleen weight, and infiltration into the central nervous system at the end of treatment (Figure 6; Table 1). Phosphorylation of STAT5 and CRKL was also attenuated in cells harvested from dasatinib-treated mice compared with vehicle-treated mice (supplemental Figure 4A). EBF1-PDFGRB is a fusion that is associated with induction failure and elevated minimal residual disease.37,38 In a model of EBF1-PDGFRB ALL (ALL14), the combination of dasatinib with continuous dexamethasone enhanced efficacy compared with either agent alone (Figure 6C-D; Table 1). Notably, the engraftment rate of this tumor was slow compared with ALL1, ALL9, and ALL14. We also observed strong synergy with dasatinib and either dexamethasone, daunorubicin, or vincristine on leukemic cells from ALL1, ALL13, and ALL14 treated ex vivo (supplemental Figure 4B).
Sensitivity of PDX models to TKIs in vivo
| Sample ID . | Kinase alteration . | Agent . | Spleen weight (mg) . | P value (vs vehicle) . | Additional genetic alterations . |
|---|---|---|---|---|---|
| ALL1 | ETV6-ABL1 | Vehicle | 321.0 ± 18.0 | NA | |
| Das | 117.4 ± 16.2 | .0014 | |||
| ALL9 | RCSD1-ABL2 | Vehicle | 479.2 ± 17.80 | IKZF1 deletion, CDKN2A/B deletion | |
| Das | 136.5 ± 29.0 | <.0001 | |||
| ALL13 | SSBP2-CSF1R | Vehicle | 430.0 ± 40.2 | IKZF1 deletion, CDKN2A/B deletion | |
| Das | 156.2 ± 12.2 | .0001 | |||
| ALL14 | EBF1-PDGFRB | Vehicle | 290.8 ± 17.5 | EBF1 deletion | |
| Das | 109.3 ± 20.1 | .011 | |||
| Dex | 111.8 ± 18.1 | NS | |||
| Dex + das | 31.5 ± 8.2 | .0009 | |||
| ALL19 | PAX5-JAK2 | Vehicle | 289.3 ± 39.9 | IKZF1 deletion | |
| Ruxo | 122.2 ± 16.8 | .0089 | |||
| Dex | 21.3 ± 2.0 | .0019 | |||
| Dex + ruxo | 17.8 ± 1 | .0013 | |||
| ALL21 | ATF7IP-JAK2 | Vehicle | 636.4 ± 23.8 | IKZF1 deletion, CDKN2A/B deletion, BTLA-CD200 deletion | |
| Ruxo | 99.6 ± 9.2 | <.0001 | |||
| Dex | 34.6 ± 3.0 | <.0001 | |||
| Dex + ruxo | 17.0 ± 1.2 | <.0001 |
| Sample ID . | Kinase alteration . | Agent . | Spleen weight (mg) . | P value (vs vehicle) . | Additional genetic alterations . |
|---|---|---|---|---|---|
| ALL1 | ETV6-ABL1 | Vehicle | 321.0 ± 18.0 | NA | |
| Das | 117.4 ± 16.2 | .0014 | |||
| ALL9 | RCSD1-ABL2 | Vehicle | 479.2 ± 17.80 | IKZF1 deletion, CDKN2A/B deletion | |
| Das | 136.5 ± 29.0 | <.0001 | |||
| ALL13 | SSBP2-CSF1R | Vehicle | 430.0 ± 40.2 | IKZF1 deletion, CDKN2A/B deletion | |
| Das | 156.2 ± 12.2 | .0001 | |||
| ALL14 | EBF1-PDGFRB | Vehicle | 290.8 ± 17.5 | EBF1 deletion | |
| Das | 109.3 ± 20.1 | .011 | |||
| Dex | 111.8 ± 18.1 | NS | |||
| Dex + das | 31.5 ± 8.2 | .0009 | |||
| ALL19 | PAX5-JAK2 | Vehicle | 289.3 ± 39.9 | IKZF1 deletion | |
| Ruxo | 122.2 ± 16.8 | .0089 | |||
| Dex | 21.3 ± 2.0 | .0019 | |||
| Dex + ruxo | 17.8 ± 1 | .0013 | |||
| ALL21 | ATF7IP-JAK2 | Vehicle | 636.4 ± 23.8 | IKZF1 deletion, CDKN2A/B deletion, BTLA-CD200 deletion | |
| Ruxo | 99.6 ± 9.2 | <.0001 | |||
| Dex | 34.6 ± 3.0 | <.0001 | |||
| Dex + ruxo | 17.0 ± 1.2 | <.0001 |
Spleen weight measured by mean ± SD (n = 5). P value calculated for analysis of slope by linear regression and unpaired 2-way t test comparing vehicle with dasatinib (ALL1, ALL9, ALL13), or 1-way ANOVA with Tukey posttest for multiple comparisons (ALL14, ALL19, ALL21). Additional genetic alterations determined by single nucleotide polymorphism profiling for recurrent copy number alterations in ALL.
Das, dasatinib; Dex, dexamethasone; NA, not available; NS, not significant; Ruxo, ruxolitinib.
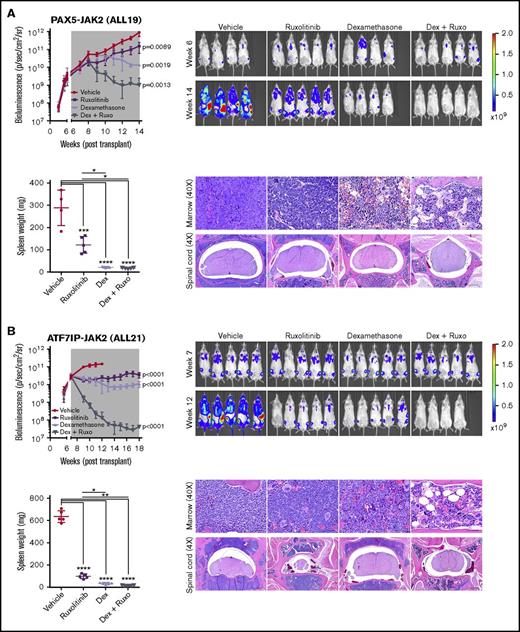
To enable more sensitive monitoring of leukemic burden in vivo, we introduced a lentiviral construct expressing yellow fluorescent protein and luciferase into 2 established PDX models harboring PAX5-JAK2 and ATF7IP-JAK2 (ALL19 and ALL21). Following engraftment, mice were randomly assigned to vehicle, ruxolitinib, dexamethasone, or a combination of ruxolitinib and dexamethasone for 5 to 7 weeks until vehicle-treated mice became moribund. Enhanced efficacy was observed with the combination of ruxolitinib and dexamethasone in both models, as evidenced by reduction of leukemic burden below pretreatment levels, lower circulating hCD45+/CD19+ blasts, and decreased infiltration into organs (Figure 7; supplemental Figure 5; Table 1). Decreased phosphorylation of STAT5 was observed in leukemic cells harvested from mice treated with ruxolitinib to a similar degree to that observed in leukemic cells harvested from vehicle-treated mice and treated with ruxolitinib ex vivo (supplemental Figure 6A). Furthermore, ruxolitinib synergized with dexamethasone in reducing the viability of JAK2-rearranged leukemic cells treated ex vivo (supplemental Figure 6B).
Superior efficacy of ruxolitinib combined with dexamethasone in JAK2-rearranged ALL. NSG mice were injected with primary leukemic cells (A) ALL19 or (B) ALL21. When engraftment reached 1010 p/sec per cm2/steridian on bioluminescent imaging, animals (n = 5) were treated with vehicle, ruxolitinib (provided as chow), dexamethasone (4 mg/mL ad libitum), or ruxolitinib and dexamethasone combined. Treatment length is indicated by the gray shaded area; animals were euthanized at the last time point. Analysis of slope was measured by linear regression and 1-way ANOVA with Tukey posttest for multiple comparisons. Spleen weights were measured at the time of euthanasia. Each point represents the mean ± SD (n = 5). Representative H&E staining of bone marrow and spinal cord of treated mice are shown. *P < .05, **P < .01, ***P < .001, ****P < .0001. Ruxo, ruxolitinib.
Superior efficacy of ruxolitinib combined with dexamethasone in JAK2-rearranged ALL. NSG mice were injected with primary leukemic cells (A) ALL19 or (B) ALL21. When engraftment reached 1010 p/sec per cm2/steridian on bioluminescent imaging, animals (n = 5) were treated with vehicle, ruxolitinib (provided as chow), dexamethasone (4 mg/mL ad libitum), or ruxolitinib and dexamethasone combined. Treatment length is indicated by the gray shaded area; animals were euthanized at the last time point. Analysis of slope was measured by linear regression and 1-way ANOVA with Tukey posttest for multiple comparisons. Spleen weights were measured at the time of euthanasia. Each point represents the mean ± SD (n = 5). Representative H&E staining of bone marrow and spinal cord of treated mice are shown. *P < .05, **P < .01, ***P < .001, ****P < .0001. Ruxo, ruxolitinib.
Another group of agents that may be efficacious in combination with TKI therapy in Ph-like ALL is the BH3 mimetic BCL-2-inhibitors (eg, venetoclax), which have demonstrated impressive single-agent activity in KMT2A-rearrranged ALL,39,40 and synergize with dasatinib in BCR-ABL1 ALL41 and JAKi in JAK-rearranged ALL.42 We observed significant synergy of venetoclax with dasatinib or ruxolitinib in leukemic cells treated ex vivo (supplemental Figure 7).
Discussion
The successful inhibition of the oncogenic tyrosine kinase activity of BCR-ABL1 with ABLi in patients with chronic myeloid leukemia43 and ALL44-46 paved the way for the identification and validation of new therapeutic targets in hematologic malignancies. Ph-like ALL is characterized by genetic alterations that activate tyrosine kinase or cytokine receptor signaling.3,4 Although kinase alterations are identified in both standard and high-risk ALL,7,47 high-risk features including treatment failure and relapse are enriched in patients with Ph-like ALL compared with non–Ph-like ALL patients; thus, new therapeutic strategies are required to improve the outcome of these patients. In this study, we provide a comprehensive assessment of the function and targetability of kinase alterations identified in Ph-like ALL to guide the development of clinical trials assessing the efficacy of TKI therapy. Comprehensive genomic analysis of Ph-like ALL patients in large cohorts of standard-risk ALL are ongoing; therefore, the clinical utility of TKIs in this patient population is unknown. However, our recent data from the St. Jude Total Therapy XV study, which has a higher proportion of ALL cases initially classified as standard-risk compared with our studies from the Children Oncology Group and adult groups, indicate that, although overall survival of Ph-like ALL cases is not inferior to non–Ph-like B-ALL cases, such cases commonly exhibit inferior response to therapy requiring treatment intensification. Moreover, such poorly responsive cases frequently have kinase-activating lesions predicted to respond to TKI therapy.47
Using cytokine-dependent Ba/F3 cells, we demonstrate that Ph-like ALL kinase fusions confer the ability to survive and grow independently of cytokine through the activation of several signaling pathways, including JAK-STAT, CRKL (ABL1, ABL2), PI3K, and MAPK. Although rearrangements of ABL1 have been described extensively in B- and T-ALL,48,49 rearrangements of ABL2, a homolog of ABL1, have rarely been identified in hematologic malignancies. Furthermore, CSF1R regulates the differentiation of macrophages and is not normally expressed in lymphocytes. Using gene expression profiling of cells obtained during normal mouse and human hematopoietic development, we demonstrate the 5′ fusion partner is commonly expressed and hijacks the 3′ kinase fusion partner to result in abnormal expression of a constitutively active kinase or cytokine receptor in leukemia cells. We also show for the first time that ABL2 and CSF1R kinase fusions are leukemogenic; their introduction into primary mouse Arf−/− pre-B cells induces the rapid development of B-ALL that recapitulates human disease. Furthermore, the presence of IKZF1 alteration accelerates progression of disease, consistent with our prior data in BCR-ABL leukemia demonstrating that loss of Ikaros function results in derepression of stem cell and adhesion pathways, leading to an aggressive tumor phenotype.15 As approximately 75% of Ph-like ALL patients harbor alterations in IKZF1, and approximately 50% of patients harbor alterations in both B-lymphoid development and the tumor suppressor CDKN2A/B, this combination of genetic features with a kinase alteration faithfully recapitulates the human disease.3,4
We provide comprehensive cell-based in vitro sensitivities for each kinase alteration against a panel of TKIs. As predicted, the ABL1 and ABL2 fusions were grouped together, followed by the type III RTKs CSF1R, FLT3, and PDGFRB. The uniform sensitivity of these kinases to dasatinib prompted further investigation of this agent in PDX models of Ph-like ALL. Single-agent treatment with dasatinib in leukemic cells harboring fusion of ABL1, ABL2, CSF1R, or PDGFRB resulted in a cytostatic effect, whereas combination with dexamethasone reduced leukemic burden to undetectable levels. These data suggest that addition of ABLi should be tested in Ph-like ALL patients harboring ABL-class fusions, and is supported by anecdotal reports demonstrating exceptional response of Ph-like ALL patients with ABL-class fusions to targeted therapies.3,37,38,50
Another major group of kinase alterations in Ph-like ALL are those that collectively activate JAK-family kinases. We tested a range of JAK inhibitors with listed specificities for each JAK kinase and identified the JAK1/JAK2 inhibitor, ruxolitinib, as the most potent agent against all JAK alterations except those involving TYK2. Thus, specific inhibitors will need to be investigated for targeting TYK2 fusions. Our in vivo PDX studies were focused on cases with JAK2 fusion. Similar to ABLi, we observed cytostatic effects with ruxolitinib monotherapy that was enhanced significantly with the addition of dexamethasone. These data and previous reports demonstrating effective inhibition of JAK-STAT–activating alterations to JAK inhibition13,14 suggest addition of ruxolitinib to chemotherapy should be tested in patients harboring these alterations. Other interesting targeted agents identified in this screen include the compound JAK3i, an irreversible acrylamide-based selective JAK3 inhibitor36 that showed exquisite potency and sensitivity toward Ba/F3 cells expressing JAK3 p.Val670Ala and MYH9-IL2RB. The similar in vitro efficacy and inhibition of pSTAT5 by JAK3i in BaF3-MYH9-IL2RB cells provides insight that this cytokine receptor rearrangement may signal through the JAK3-STAT5 pathway, similar to wild-type IL2RB.51,52 Compared with crizotinib, the TRK/CSF1R-specific inhibitor, PLX7486, was 10-fold more potent against the ETV6-NTRK3 fusion. Both JAK3i and PLX7486 are interesting agents that require in vivo studies to assess clinical potential.
A consistent finding from these studies is that the use of TKIs as single-agent treatment in ALL is of limited efficacy in eradicating disease, and that the combination of multiple agents is required for superior efficacy, as has been observed in BCR-ABL1 ALL in contradistinction to chronic myeloid leukemia, in which TKI monotherapy is highly efficacious.43 Similar to our previous studies of BCR-ABL1 and EPOR-rearranged ALL,15,17 we demonstrate that the combination of ABLi or JAKi with dexamethasone enhances efficacy and reduces leukemic burden to undetectable levels. This has also been observed in elderly BCR-ABL1 ALL patients in whom induction therapy with inhibitor and steroids alone induced complete hematologic remission in >90% of patients studied.53,54 An interesting body of work by Tasian et al has established that the activation of PI3K signaling is an important downstream effector of kinase signaling in Ph-like ALL, particularly in leukemias harboring rearrangement of CRLF2.14,55 They also demonstrate a clear dependence of Ph-like ALL leukemic cells on PI3K signaling, with enhanced killing and survival using JAKi in combination with the dual PI3K/mTOR inhibitor, gedatolisib. This is one of the first studies to explore the use of multitargeted therapy in the absence of chemotherapy. We provide evidence in vitro that the combination of dasatinib or ruxolitinib with BCL2 inhibition is synergistic. Future studies assessing the efficacy of this combination in vivo are required to assess the feasibility of this treatment approach. Lastly, agents such as retinoids and focal adhesion kinase inhibitors that mitigate aberrant activation of integrin signaling cascades precipitated by IKZF1 alterations in BCR-ABL1 ALL could also be explored in Ph-like ALL.15,56
Our findings demonstrate that, despite the large number of individual kinase alterations identified in Ph-like ALL, the majority converge on a limited number of pathways that can be targeted effectively in vivo using ABLi or JAKi in combination with dexamethasone. These data provide the rationale for testing the efficacy of dasatinib added to a chemotherapy backbone in patients harboring ABL1-class alterations and has been implemented by both the Children’s Oncology Group (www.clinicaltrials.gov: #NCT02883049) and St. Jude Children’s Research Hospital (www.clinicaltrials.gov: #NCT03117751 [Total Therapy XVII]). The efficacy of ruxolitinib in addition to chemotherapy is being assessed in patients harboring JAK-STAT alterations in the Children’s Oncology Group trial AALL1521 (www.clinicaltrials.gov: #NCT02723994) and Total Therapy XVII.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the staff of the Biorepository, the Flow Cytometry and Cell Sorting Core Facility, and the Small Animal Imaging Core at St. Jude Children’s Research Hospital. The MSCV-ETV6-NTRK3-ires-Cherry construct was provided by Suzanne Baker at St. Jude Children’s Research Hospital. Ba/F3 cells expressing FLT3 ITD were obtained from Sharyn Baker, formerly of St. Jude Children’s Research Hospital. The authors also thank Incyte Corporation for providing ruxolitinib and Plexxikon for providing PLX7486 and PLX3397.
This work was supported in part by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital, an Alex’s Lemonade Stand Foundation Young Investigator Award (K.G.R.), Leukemia and Lymphoma Society Special Fellow Award (K.G.R.), American Society of Hematology Scholar Award (K.G.R.), Stand Up to Cancer Innovative Research Grant, St. Baldrick’s Foundation Scholar Award (C.G.M.), St. Baldrick’s Consortium Award (S.P.H.), Leukemia and Lymphoma Society Specialized Center of Research grant (C.G.M. and S.P.H.), and the National Institutes of Health, National Cancer Institute (grants CA21765 [St. Jude Cancer Center Support Grant] and U10 CA 180886 and U10 CA 180899 [Children’s Oncology Group]). S.P.H. is the Jeffrey E. Perelman Distinguished Chair in the Department of Pediatrics, Children's Hospital of Philadelphia.
Authorship
Contribution: K.G.R. designed, directed, and performed research, analyzed data, and wrote the manuscript; Y.-L.Y., D.P.-T., W.L., J.K.F., and K.D. performed research and analyzed data; Z.G. and L.J.J. analyzed data; J.T. and M.L.L. provided key reagents; T.C. contributed to research design; S.P.H. and C.G.M. contributed to research design and edited the manuscript; and all authors critically reviewed the manuscript before submission.
Conflict-of-interest disclosure: S.P.H. has received consulting fees from Novartis and honoraria from Amgen. C.G.M. has received consulting fees and honoraria from Incyte and Amgen. The Ph-like gene expression classifier used in this work is covered in part by pending US application 20140322166 (inventors: S.P.H., C.G.M., and K.G.R.; applicants: STC.UNM, St. Jude Children’s Research Hospital, and The Children’s Hospital of Philadelphia on behalf of Children’s Oncology Group).
The current affiliation for Y.-L.Y. is Department of Laboratory Medicine, National Taiwan University Hospital, College of Medicine, National Taiwan University, Taipei, Taiwan.
Correspondence: Charles G. Mullighan, Department of Pathology, 262 Danny Thomas Pl, St. Jude Children’s Research Hospital, Memphis, TN 38105; e-mail: charles.mullighan@stjude.org.
References
Author notes
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE38463).