Key Points
TLR1 is upregulated on primitive AML cells.
Agonistic targeting of TLR1/TLR2 induces apoptosis and differentiation of primitive AML cells in vivo.
Abstract
Acute myeloid leukemia (AML) is associated with poor survival, and there is a strong need to identify disease vulnerabilities that might reveal new treatment opportunities. Here, we found that Toll-like receptor 1 (TLR1) and TLR2 are upregulated on primary AML CD34+CD38− cells relative to corresponding normal bone marrow cells. Activating the TLR1/TLR2 complex by the agonist Pam3CSK4 in MLL-AF9-driven human AML resulted in induction of apoptosis by p38 MAPK-dependent activation of Caspase 3 and myeloid differentiation in a NFκB-dependent manner. By using murine Trp53−/−MLL-AF9 AML cells, we demonstrate that p53 is dispensable for Pam3CSK4-induced apoptosis and differentiation. Moreover, murine AML1-ETO9a-driven AML cells also were forced into apoptosis and differentiation on TLR1/TLR2 activation, demonstrating that the antileukemic effects observed were not confined to MLL-rearranged AML. We further evaluated whether Pam3CSK4 would exhibit selective antileukemic effects. Ex vivo Pam3CSK4 treatment inhibited murine and human leukemia-initiating cells, whereas murine normal hematopoietic stem and progenitor cells (HSPCs) were relatively less affected. Consistent with these findings, primary human AML cells across several genetic subtypes of AML were more vulnerable for TLR1/TLR2 activation relative to normal human HSPCs. In the MLL-AF9 AML mouse model, treatment with Pam3CSK4 provided proof of concept for in vivo therapeutic efficacy. Our results demonstrate that TLR1 and TLR2 are upregulated on primitive AML cells and that agonistic targeting of TLR1/TLR2 forces AML cells into apoptosis by p38 MAPK-dependent activation of Caspase 3, and differentiation by activating NFκB, thus revealing a new putative strategy for therapeutically targeting AML cells.
Introduction
Acute myeloid leukemia (AML) is characterized by a clonal expansion of myeloid leukemia blasts with impaired differentiation that accumulate in the bone marrow (BM).1 Because the prognosis for patients with AML is generally poor (the 5-year overall survival is around 20% in patients older than 60 years),2 demand is strong for new types of therapies. One of the major obstacles in successfully treating AML is to efficiently target the leukemic stem cells (LSCs), a self-renewing immature cell population with leukemia-initiating capacity, which often evades standard chemotherapy treatments, leading to disease relapse.1,3,4 Hence, the development of new therapeutic strategies that more efficiently target the LSCs is warranted. One approach to targeting LSCs is to identify cell surface proteins upregulated on these cells. Antibodies targeting cell surface proteins such as CD47,2,5 CD123,6 IL1RAP,7 CLL-1,8 and TIM-3,9 upregulated on LSCs, have shown therapeutic efficacy in preclinical models by blocking their targets and/or stimulating the immune system to attack the leukemic cells.
Another approach to targeting AML stem cells is differentiation-based therapies, successfully used in the acute promyelocytic leukemia subtype, in which all-trans retinoic acid, a nonchemotherapeutic drug, has had a significant effect on survival.10,11 All-trans retinoic acid acts by forcing leukemic promyelocytes into granulocytic differentiation11,12 and apoptosis.13 Recent advances have been made to differentiate other AML subtypes, such as isocitrate dehydrogenase 1/2 mutated AML.14-16 These findings suggest that differentiation-based therapies may reach clinical utility also in other subtypes of AML.
Activation of Toll-like receptor 1 (TLR1), which is a co-receptor to TLR2,17,18 results in differentiation of normal myeloid progenitor cells.19 The TLR1/TLR2 complex is a part of the first line of defense against bacterial infections by binding triacylated lipopeptides.17,20 TLR1 mRNA levels are upregulated in CD34+ BM cells from patients with myelodysplastic syndrome, and TLR8 activation of AML cells has recently been shown to result in cell differentiation.21-23 However, whether TLR1 is upregulated in AML and whether activation of the TLR1/TLR2 complex induces differentiation of AML cells has not been previously explored.
Here, we report that TLR1 and TLR2 are upregulated on immature AML cells and that TLR1/TLR2 activation induces differentiation of AML cells by activating NFκB-signaling and apoptosis via p38 MAPK and Caspase 3. These findings suggest that activation of TLR1/TLR2 provides a novel strategy to therapeutically target AML cells in vivo, findings that may translate into new therapeutic opportunities in AML.
Materials and methods
Murine leukemia models
All animal experiments were approved by the local ethics committee in Lund, Sweden. Mouse MLL-AF9 leukemias in a C57BL/6 dsRed transgenic background (6051; The Jackson Laboratory, Bar Harbor, ME), and Trp53−/− background (002101; The Jackson Laboratory), were previously generated by retroviral expression of MLL-AF9 in murine hematopoietic stem and progenitor cells (HSPCs).24,25 Similarly, mouse AML1-ETO9a leukemias were previously generated by retroviral expression of AML1-ETO9a along with EGFP in HSPCs.26,27 Leukemia cells were propagated by transplantation into sublethally irradiated (600 cGy) C57BL/6 mice recipients. All experiments involving murine MLL-AF9 leukemia cells were performed using tertiary or quaternary transplanted leukemia cells and murine AML1-ETO9a leukemia cells using secondary or tertiary transplanted leukemia cells. On disease development, BM cells from femurs and tibias of leukemic mice were isolated by crushing the bones, filtering, and lysis of red blood cells using NH4Cl (StemCell Technologies, Vancouver, Canada). To enrich for LSCs in both models, c-Kit+ BM cells were purified using midi MACS columns and anti-CD117 microbeads according to the manufacturer’s instructions (Miltenyi Biotec, Bergisch Gladbach, Germany).
Human cells
All BM and peripheral blood samples from patients with AML and from healthy donors were obtained after informed consent according to protocols approved by the regional ethics committee in Lund, Sweden. Mononuclear cells (MNCs) were separated using Lymphoprep (Axis-Shield PoC AS, Dundee, United Kingdom). To enrich for primitive cells, CD34+ cells from normal BM samples were isolated using a CD34+ cell isolation kit, according to the manufacturer’s instructions (Miltenyi Biotec).
Flow cytometric analysis
Flow cytometric analyses were performed in either a FACSCanto II flow cytometer or a LSRFortessa instrument, and flow cytometric cell sorting was performed in a FACSAria IIU or FACSAria Fusion cell sorter (all from BD Biosciences, San Jose, CA). For details on antibody staining, see supplemental Methods.
Cultures of leukemia and normal BM cells
Primary human CD34+CD38− cells were cultured in S7 medium as previously described.28 MA9 cells were previously generated by retroviral expression of the MLL-AF9 fusion gene in primitive cord blood cells.29 For further details on culturing conditions, see supplemental Methods. Pam3CSK4 (InvivoGen, San Diego, CA) was added to the culture medium at a final concentration of 1 μg/mL unless otherwise stated. CU-T12-930 (Tocris Bioscience, Bristol, United Kingdom) was used at 1 μM.
Cell proliferation and apoptosis
To determine the number of cells in culture, flow cytometric analysis with CountBright absolute counting beads (BD Biosciences) was performed. The amount of apoptotic cells after culturing cells with Pam3CSK4 for 24, 48, or 72 hours, respectively, was assessed by staining the cells in 1× binding buffer (BD Biosciences) with APC-labeled Annexin V and 7-amino-actinomycin (both from BD Biosciences). For the apoptosis analysis of murine dsRed+ leukemia cells, 4′,6-diamidino-2-phenylindole (DAPI; BioLegend, San Diego, CA) was used instead of 7-amino-actinomycin. In TLR1- and TLR2-antibody-blocking experiments, the human leukemia cells were stimulated with 10 ng/mL Pam3CSK4. For more information on antibody-blocking experiments, see supplemental Methods. Staining of cleaved (active) Caspase 3 was performed using a Caspase 3-PE mAb (Cell signaling Technologies, Danvers, MA) and measured by flow cytometry in fixed and permeabilized cells, performed as described for the phospho-flow cytometric analysis in supplemental Methods.
Small molecule inhibition
To inhibit NFκB phosphorylation, the leukemia cells were preincubated with 1 or 3 μM of the IκB kinase inhibitor TPCA1 (Sigma-Aldrich). To inhibit p38 MAPK phosphorylation, 1 or 10 μM of the p38 MAPK inhibitor SB203580 (Sigma-Aldrich) was used.
Colony assay
MA9 cells were cultured for 3 days with Pam3CSK4 and then seeded into methylcellulose medium (MethoCult H4434; StemCell Technologies). The number of colonies was scored after 14 days of incubation at 37°C.
Ex vivo Pam3CSK4 treatment of leukemia cells or LSK cells
Murine c-Kit+MLL-AF9 leukemia cells were treated with Pam3CSK4 for 3 days and then injected into sublethally irradiated (600 cGy) recipient mice. Blood sampling was performed 2 weeks posttransplantation, and the percentage of dsRed+ leukemic cells was determined using flow cytometric analysis. Five million human MA9 cells were treated with Pam3CSK4 for 3 days and then injected into sublethally irradiated (250 cGy) NOD scid γ (NSG; The Jackson Laboratory) mice. Blood samplings of the NSG mice were performed 4 weeks posttransplantation, and the experiment was terminated 16 weeks posttransplantation. The percentage of leukemia (human CD45+) cells was determined using flow cytometry. Sorted Linage−Sca1+c-Kit+ (LSK) cells from B6SJL (CD45.1+) and B6SJL-C57BL/6 (CD45.1+CD45.2+) mice were cultured in SFEM supplemented with 100 ng/mL murine thrombopoietin and 20 ng/mL murine stem cell factor (SCF) for 3 days. Pam3CSK4 was added to cultures with CD45.1+CD45.2+ cells. After culture, CD45.1+CD45.2+ and CD45.1+ (control) cells were mixed and injected into lethally irradiated (900 cGy) recipient (CD45.2+) mice along with 200 000 support cells (CD45.2+). At 16 weeks posttransplantation, BM cells were harvested as described earlier for leukemia cells and analyzed by flow cytometry.
RNA sequencing analysis
MA9 cells were stimulated with Pam3CSK4 for 3 and 24 hours. For details on RNA extraction, library preparations, sequencing, and analysis, see supplemental Methods. Raw data and normalized gene expression data are available in the Gene Expression Omnibus database under accession no. GSE92744.
Normalized AML patient RNA sequencing data (reads per kilobase million) were downloaded from the TCGA database.31 Qlucore Omics Explorer 3.0 (Qlucore, Lund, Sweden) was used to analyze the data.
Statistics
Kaplan-Meier curves were generated using Prism 5 software (GraphPad Software, La Jolla, CA). In all figures, the mean values and standard deviations (SDs) are shown. P values were calculated using the Student t test, the Mann-Whitney U test (for TLR1 expression in primary human cells only), or Mantel-Cox test (for Kaplan-Meier curves only). Correlation between TLR1 and TLR2 mRNA expression was calculated using Spearman’s rank correlation. Significance is indicated with asterisks (*P < .05; **P < .01; ***P < .001; ****P < .0001).
Results
TLR1 and TLR2 are upregulated on immature AML patient cells
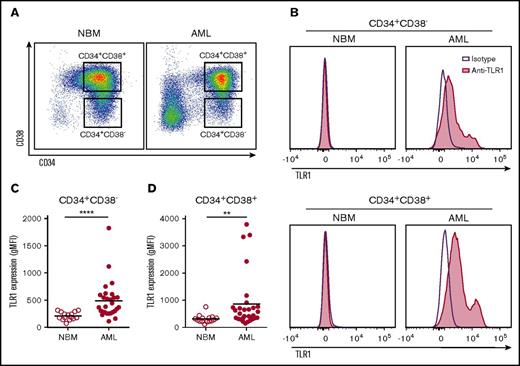
In a search for upregulated cell surface proteins on immature AML cells, we measured TLR1 expression on cells from a consecutive series of 28 patients with AML (supplemental Table 1). Within the AML CD34+CD38− cell compartment (Figure 1A), enriched for AML stem cells,32 TLR1 was upregulated in about 50% of the samples relative to corresponding normal BM cells, which were devoid of TLR1 expression (Figure 1B-C). Similar upregulation of TLR1 was observed in AML CD34+CD38+ cells (Figure 1A-B,D) and bulk MNCs (supplemental Figure 1A). No correlation between TLR1 expression and genetic (karyotype or mutations in FLT3 or NPM1) or clinical subgroups (FAB-subgroups or percentage blasts in the BM) of patients with AML could be found (supplemental Table 1). However, in RNA sequencing data of AML from the TCGA,31 we found a correlation between upregulated TLR1 mRNA expression and patients with AML harboring a CBFB-MYH11 fusion gene (fold-change, 2.37; P < .001; supplemental Table 2). We also studied the expression levels of TLR2, which is the signaling partner for TLR1. A moderate correlation between TLR1 and TLR2 mRNA expression was found in the TCGA database (rs = 0.445; P < .001; supplemental Figure 1B). Similar to TLR1, flow cytometric analysis revealed that TLR2 was expressed in CD34+CD38− cells from 5 of 8 AML patient samples analyzed; corresponding normal BM cells lacked TLR2 expression (supplemental Figure 1C-D).
TLR1 was upregulated on about half of the primary human AML samples analyzed. (A) FACS dot plots showing gating of CD34+CD38− and CD34+CD38+ in representative CD34-enriched normal bone marrow (NBM) and primary AML patient samples. (B) Histograms depicting TLR1 expression (purple line: isotype control antibody, red: anti-TLR1 antibody) within CD34+CD38− (top histograms) and CD34+CD38+ (bottom histograms) cells from representative NBM and TLR1-high AML samples, respectively. Geometric mean fluorescence intensity of TLR1 expression in NBM (n = 15) and AML (n = 28) (C) CD34+CD38− and (D) CD34+CD38+ cells. Each dot represents a sample from 1 patient, and horizontal lines indicate mean geometric mean fluorescence intensity for all samples within groups. **P < .01; ****P < .0001.
TLR1 was upregulated on about half of the primary human AML samples analyzed. (A) FACS dot plots showing gating of CD34+CD38− and CD34+CD38+ in representative CD34-enriched normal bone marrow (NBM) and primary AML patient samples. (B) Histograms depicting TLR1 expression (purple line: isotype control antibody, red: anti-TLR1 antibody) within CD34+CD38− (top histograms) and CD34+CD38+ (bottom histograms) cells from representative NBM and TLR1-high AML samples, respectively. Geometric mean fluorescence intensity of TLR1 expression in NBM (n = 15) and AML (n = 28) (C) CD34+CD38− and (D) CD34+CD38+ cells. Each dot represents a sample from 1 patient, and horizontal lines indicate mean geometric mean fluorescence intensity for all samples within groups. **P < .01; ****P < .0001.
TLR1/TLR2 activation induces myeloid differentiation of AML cells
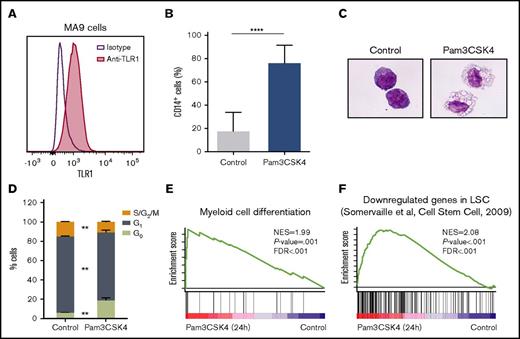
Having found that TLR1 and TLR2 are upregulated on immature AML cells, we next explored whether agonistic targeting of the TLR1/TLR2 receptor complex on AML cells using Pam3CSK4, a synthetic lipopeptide that mimics the natural ligand of TLR1/TLR2,18,33 would have an effect on AML cells. TLR1 and TLR2 were expressed on MA9 cells, a cytokine-dependent human AML cell line driven by enforced MLL-AF9 expression (Figure 2A; supplemental Figure 2A).29 Pam3CSK4 treatment of the MA9 cells resulted in a strong increase in CD14 expression consistent with myeloid differentiation, accompanied by a weak downregulation of TLR1 (Figure 2B; supplemental Figure 2B). Morphological assessments confirmed the outgrowth of macrophage-like cells (Figure 2C). The differentiation was also accompanied by a cell cycle arrest (Figure 2D; supplemental Figure 2C). Consistent with these observations, RNA sequencing of Pam3CSK4-treated MA9 cells revealed enrichment of myeloid cell differentiation signatures, as assessed by gene set enrichment analysis (GSEA) (Figure 2E; supplemental Table 3). Moreover, we confirmed that Pam3CSK4 induces differentiation of MA9 cells in a TLR1/TLR2-dependent manner by blocking either TLR1 or TLR2, using neutralizing antibodies (supplemental Figure 2D).34,35 In line with the differentiation induced by TLR1/TLR2 activation, GSEA also revealed an enrichment of genes that are negatively correlated with LSC frequency in the Pam3CSK4 signature (Figure 2F; supplemental Table 3).
TLR1/TLR2 activation leads to differentiation of AML cells. (A) TLR1 expression on human MA9 leukemia cells. Histogram showing MA9 cells stained for TLR1 (red) and isotype control (purple line) using flow cytometry. (B-D) MA9 cells were treated with or without (control) Pam3CSK4 for 3 days. (B) Percentage of CD14+ cells measured by flow cytometry (n = 3). (C) May-Grünwald-Giemsa–stained cytospin slides (40× magnification). (D) Cell cycle status of MA9 cells from 1 representative experiment of 3. (E-F) In MA9 cells treated with Pam3CSK4 for 24 h, GSEA identified an enriched (E) myeloid cell differentiation signature and (F) a LSCs down signature; that is, a signature of genes downregulated in LSCs (AML cell populations with high versus low LSCs frequency). Mean values and SDs are shown. **P < .01; ****P < .0001.FDR, false discovery rate; NES, normalized enrichment score.
TLR1/TLR2 activation leads to differentiation of AML cells. (A) TLR1 expression on human MA9 leukemia cells. Histogram showing MA9 cells stained for TLR1 (red) and isotype control (purple line) using flow cytometry. (B-D) MA9 cells were treated with or without (control) Pam3CSK4 for 3 days. (B) Percentage of CD14+ cells measured by flow cytometry (n = 3). (C) May-Grünwald-Giemsa–stained cytospin slides (40× magnification). (D) Cell cycle status of MA9 cells from 1 representative experiment of 3. (E-F) In MA9 cells treated with Pam3CSK4 for 24 h, GSEA identified an enriched (E) myeloid cell differentiation signature and (F) a LSCs down signature; that is, a signature of genes downregulated in LSCs (AML cell populations with high versus low LSCs frequency). Mean values and SDs are shown. **P < .01; ****P < .0001.FDR, false discovery rate; NES, normalized enrichment score.
TLR1/TLR2 activation forces AML cells into apoptosis by activating Caspase 3
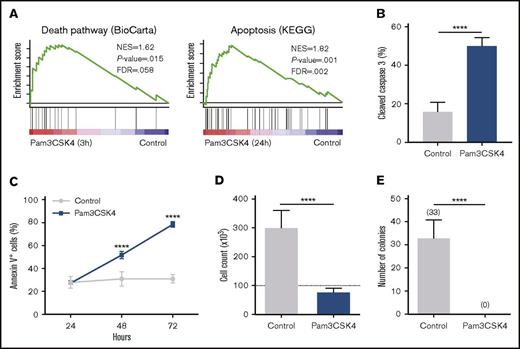
In search for additional transcriptional changes in MA9 cells on TLR1/TLR2 activation, we identified an enrichment of several apoptosis and cell death pathway signatures in the Pam3CSK4-treated cells relative to control cells, already after 3 hours of TLR1/TLR2 activation (Figure 3A; supplemental Table 3). Consistent with these observations, agonistic targeting of TLR1/TLR2 on MA9 cells resulted in Caspase 3 activation (Figure 3B) and an increase in apoptosis as assessed by Annexin V staining (Figure 3C; supplemental Figure 2E). Similar to the Pam3CSK4-induced differentiation, the TLR1 and TLR2 blocking antibodies partially inhibited Pam3CSK4-induced apoptosis of the MA9 cells and did not alter the differentiation or apoptosis status of the MA9 cells in the absence of Pam3CSK4 stimulation (supplemental Figure 2F-H). These findings show that Pam3CSK4 induces both differentiation and apoptosis of AML cells in a TLR1/TLR2-dependent manner. Moreover, TLR1/TLR2 activation inhibited cell proliferation (Figure 3D) and eradicated the colony-forming capacity of the MA9 cells (Figure 3E). Similar findings were obtained using CU-T12-9, another TLR1/TLR2 agonist, further supporting that TLR1/TLR2 activation triggers apoptosis and differentiation of AML cells (supplemental Figure 2I-J).
TLR1/TLR2 activation induces apoptosis of AML cells via Caspase 3 activation. (A) In MA9 cells treated with Pam3CSK4, GSEA revealed enriched death pathway (3 hours; left panel) and apoptosis (24 hours; right panel) signatures. (B-E) MA9 cells treated with or without (control) Pam3CSK4 for 3 days. (B) Percentage of cleaved (active) Caspase 3+ cells and (C) percentage of Annexin V+ cells over time. (D) Cell count as determined by flow cytometry, using CountBright beads. Dotted line indicates amount of cells seeded. (E) Number of colonies formed in methylcellulose after 14 days. Shown in all graphs are data from 3 independent experiments, each with 3 replicates. Mean values and SDs are shown. ****P < .0001.
TLR1/TLR2 activation induces apoptosis of AML cells via Caspase 3 activation. (A) In MA9 cells treated with Pam3CSK4, GSEA revealed enriched death pathway (3 hours; left panel) and apoptosis (24 hours; right panel) signatures. (B-E) MA9 cells treated with or without (control) Pam3CSK4 for 3 days. (B) Percentage of cleaved (active) Caspase 3+ cells and (C) percentage of Annexin V+ cells over time. (D) Cell count as determined by flow cytometry, using CountBright beads. Dotted line indicates amount of cells seeded. (E) Number of colonies formed in methylcellulose after 14 days. Shown in all graphs are data from 3 independent experiments, each with 3 replicates. Mean values and SDs are shown. ****P < .0001.
Pam3CSK4 induces differentiation via NFκB and apoptosis via p38 MAPK
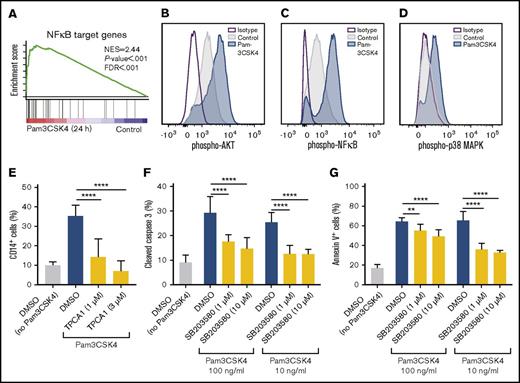
In agreement with NFκB being downstream of TLR1/TLR2 activation,35 GSEA identified an enriched NFκB signature in Pam3CSK4-treated MA9 cells relative to nontreated cells (Figure 4A; supplemental Table 3). We found that AKT, NFκB, and p38 MAPK became phosphorylated on Pam3CSK4 treatment (Figure 4B-D). By performing TLR1- and TLR2-antibody blocking experiments, we confirmed that the activation of AKT, NFκB, and p38 MAPK by Pam3CSK4 was TLR1/TLR2-dependent (supplemental Figure 3A-C).
TLR1/TLR2 activation induces differentiation of AML cells in an NFκB-dependent manner and apoptosis in a p38 MAPK-dependent manner. (A) In MA9 cells treated with Pam3CSK4 for 24 hours, GSEA identified an enriched NFκB signature relative to nontreated cells (control). (B-D) Histograms depicting phospho-flow cytometric analysis of phosphorylated (B) AKT, (C) NFκB, and (D) p38 MAPK in Pam3CSK4-treated (20 minutes) MA9 cells (blue) compared with control treated cells (gray) and isotype control (purple line). (E) Percentage of CD14+ cells after preincubation with the IκB kinase-inhibitor TPCA1 at 1 or 3 μM, or DMSO control for 10 minutes, before being treated with Pam3CSK4 (1 μg/mL) for 3 days (n = 3). (F) Percentage of cleaved (active) Caspase 3+ cells and (G) percentage of Annexin V+ cells after preincubation with the p38 MAPK inhibitor SB203580 at 1 or 10 μM, or DMSO control for 10 minutes, before being treated with Pam3CSK4 (at the indicated concentrations) for 2 days (n = 3). Mean values and SDs are shown. **P < .01; ****P < .0001.
TLR1/TLR2 activation induces differentiation of AML cells in an NFκB-dependent manner and apoptosis in a p38 MAPK-dependent manner. (A) In MA9 cells treated with Pam3CSK4 for 24 hours, GSEA identified an enriched NFκB signature relative to nontreated cells (control). (B-D) Histograms depicting phospho-flow cytometric analysis of phosphorylated (B) AKT, (C) NFκB, and (D) p38 MAPK in Pam3CSK4-treated (20 minutes) MA9 cells (blue) compared with control treated cells (gray) and isotype control (purple line). (E) Percentage of CD14+ cells after preincubation with the IκB kinase-inhibitor TPCA1 at 1 or 3 μM, or DMSO control for 10 minutes, before being treated with Pam3CSK4 (1 μg/mL) for 3 days (n = 3). (F) Percentage of cleaved (active) Caspase 3+ cells and (G) percentage of Annexin V+ cells after preincubation with the p38 MAPK inhibitor SB203580 at 1 or 10 μM, or DMSO control for 10 minutes, before being treated with Pam3CSK4 (at the indicated concentrations) for 2 days (n = 3). Mean values and SDs are shown. **P < .01; ****P < .0001.
To determine whether the Pam3CSK4-induced differentiation and apoptosis of leukemia cells is dependent on NFκB-signaling, we used TPCA1, an inhibitor of IκB kinase.36 TPCA1 blocked NFκB phosphorylation and differentiation induced by Pam3CSK4 in MA9 cells (Figure 4E; supplemental Figure 4A), demonstrating that TLR1/TLR2 activation results in differentiation of AML cells in an NFκB-dependent manner. However, TPCA1 did not block the apoptotic effects induced by Pam3CSK4, suggesting the Pam3CSK4-induced apoptosis was independent of NFκB-signaling (supplemental Figure 4B). In contrast, we found that the apoptotic effect of Pam3CSK4 was mediated via p38 MAPK signaling, using the selective p38 MAPK inhibitor SB203580, which impeded Pam3CSK4-induced Caspase 3 activation and apoptosis of MA9 cells (Figure 4F-G; supplemental Figure 4C). In addition, inhibition of p38 MAPK by SB203580 did not affect NFκB phosphorylation and only mildly suppressed the differentiation induced by Pam3CSK4 in MA9 cells (supplemental Figure 4D-E), suggesting that the p38 MAPK is more central for the Caspase 3 activation and apoptosis triggered by TLR1/TLR2 activation.
TLR1/TLR2 activation suppresses leukemia-initiating cells
We next explored whether agonistic targeting of the TLR1/TLR2 complex negatively affects leukemia-initiating cells on transplantation to irradiated hosts. To this end, we used a murine AML model driven by the MLL-AF9 fusion gene in a dsRed-transgenic background, previously used to reveal novel biology of LSCs.24,25,37,38 Serial transplantations result in reproducible engraftment of leukemia cells in BM and spleens of recipient mice.24 We found TLR1 to be expressed on leukemic granulocyte and macrophage progenitor cells, enriched for LSCs in this model (supplemental Figure 5A).39 Moreover, TLR2 was expressed on c-Kit+ leukemia cells (supplemental Figure 5B). Similar to the effects observed in the MA9 cells, TLR1/TLR2 activation resulted in apoptosis and differentiation (Figure 5A-C). In addition, a decreased cell cycling, reduced output of cells in culture, and NFκB phosphorylation was observed on TLR1/TLR2 activation (supplemental Figure 5C-E). To evaluate whether there was a correlation between TLR1 expression levels and response to Pam3CSK4, we sorted the 10% highest and 10% lowest TLR1 expressing cells and stimulated them with Pam3CSK4 for 3 days (supplemental Figure 5F). Pam3CSK4 induced similar levels of apoptosis and differentiation in the TLR1 low and high leukemia cells, suggesting that the expression level of TLR1 did not determine their sensitivity to Pam3CSK4 in this context (supplemental Figure 5G-H). To assess the effect of TLR1/TLR2 activation in leukemia-initiating cells, we treated c-Kit+MLL-AF9 leukemia cells ex vivo with Pam3CSK4 for 3 days and then transplanted the cells into sublethally irradiated mice (the Pam3CSK4 group). After 2 weeks, leukemic cells were more than 10-fold reduced in the peripheral blood of the Pam3CSK4 group compared with the control group (supplemental Figure 6A). Moreover, the Pam3CSK4 group exhibited significantly prolonged survival relative to the control mice (median, 36 vs 29 days [P = .0015; Figure 5D]; and median 47 vs 34.5 days [P = .0011; Figure 5E]). As the mice succumbed to disease and were killed, they all had a high leukemia burden in the BM and enlarged spleens, consistent with a fully developed disease (supplemental Figure 6B-C). These findings show that agonistic targeting of TLR1/TLR2 inhibits LSCs. In corresponding ex vivo treatment experiments using human MA9 cells followed by transplantations into NSG mice, in the Pam3CSK4 group, we observed lower leukemic burden in peripheral blood at 4 weeks posttransplantation and prolonged survival (median, 126 days vs not reached; P = .0049; supplemental Figure 7A-B), accompanied by reduced leukemia burden in the BM and smaller spleens (supplemental Figure 7C-D). These findings demonstrate that TLR1/TLR2 activation inhibits both murine and human leukemia initiating cells.
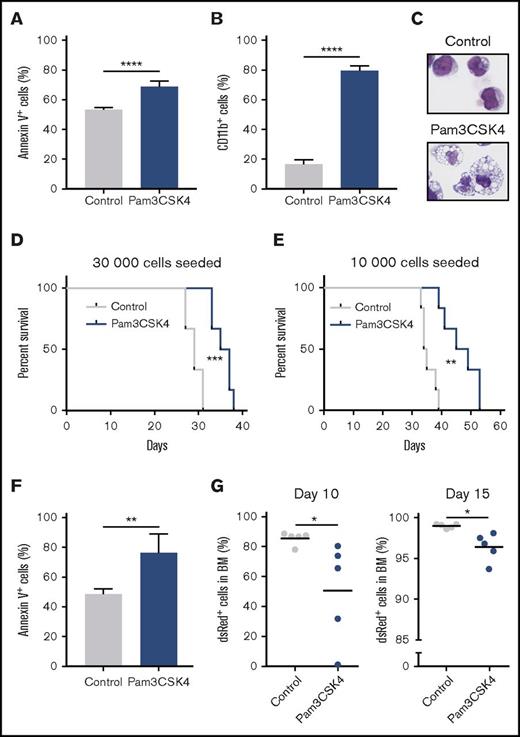
Agonistic targeting of TLR1/TLR2 inhibits leukemia-initiating cells and provides antileukemic effects in vivo. (A-E) Murine c-Kit+MLL-AF9 AML cells were cultured for 3 days with or without (control) Pam3CSK4. (A) Percentage of Annexin V+ and (B) CD11b+ cells (n = 3). (C) May-Grünwald-Giemsa–stained cytospin slides from representative samples (40× magnification). (D) 30 000 or (E) 10 000 leukemia cells were seeded in wells and cultured with Pam3CSK4 and SCF (Pam3CSK4) or only SCF (control), and then transplanted into sublethally irradiated recipient mice (6 mice per group). For the “30 000 seeded cells,” on average, 9200 control and 115 000 Pam3CSK4-treated cells were injected per mouse. For the “10 000 seeded cells”: 1100 control and 27 000 Pam3CSK4-treated cells were injected per mouse. Kaplan-Meier curves showing overall survival of the mice. (F-G) 100 000 c-Kit+MLL-AF9 AML cells were transplanted into sublethally irradiated recipient mice (5 mice per group in 2 independent experiments) and treated with Pam3CSK4 (100 μg/dose) or PBS control by intraperitoneal injections 3 times per week starting 1 day posttransplantation. (F) Percentage of Annexin V+ cells in BM at day 10 posttransplantation. (G) Percentage of dsRed+ leukemia cells in BM at day 10 (left) or day 15 (right) posttransplantation. Mean values and standard deviations are presented. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Agonistic targeting of TLR1/TLR2 inhibits leukemia-initiating cells and provides antileukemic effects in vivo. (A-E) Murine c-Kit+MLL-AF9 AML cells were cultured for 3 days with or without (control) Pam3CSK4. (A) Percentage of Annexin V+ and (B) CD11b+ cells (n = 3). (C) May-Grünwald-Giemsa–stained cytospin slides from representative samples (40× magnification). (D) 30 000 or (E) 10 000 leukemia cells were seeded in wells and cultured with Pam3CSK4 and SCF (Pam3CSK4) or only SCF (control), and then transplanted into sublethally irradiated recipient mice (6 mice per group). For the “30 000 seeded cells,” on average, 9200 control and 115 000 Pam3CSK4-treated cells were injected per mouse. For the “10 000 seeded cells”: 1100 control and 27 000 Pam3CSK4-treated cells were injected per mouse. Kaplan-Meier curves showing overall survival of the mice. (F-G) 100 000 c-Kit+MLL-AF9 AML cells were transplanted into sublethally irradiated recipient mice (5 mice per group in 2 independent experiments) and treated with Pam3CSK4 (100 μg/dose) or PBS control by intraperitoneal injections 3 times per week starting 1 day posttransplantation. (F) Percentage of Annexin V+ cells in BM at day 10 posttransplantation. (G) Percentage of dsRed+ leukemia cells in BM at day 10 (left) or day 15 (right) posttransplantation. Mean values and standard deviations are presented. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Treating mice with Pam3CSK4 shows in vivo antileukemic efficacy
To explore in vivo antileukemic efficacy of Pam3CSK4 in leukemic mice, we transplanted MLL-AF9 murine leukemia cells into sublethally irradiated recipient mice and treated them with Pam3CSK4 3 times per week. Consistent with the in vitro data, Pam3CSK4 treatment induced apoptosis and myeloid differentiation of the leukemic cells also in vivo, accompanied by a lower leukemic burden in the BM (Figure 5F-G; supplemental Figure 8A-C). However, consecutive Pam3CSK4 treatments were not well tolerated by the mice; thus, we did not perform survival experiments.
TLR1/TLR2 activation triggers apoptosis and differentiation of Trp53−/−MLL-AF9, and AML1-ETO9a AML cells
To address whether Pam3CSK4-induced apoptosis is mediated by p53, a well-known regulator of apoptosis,40 we used murine Trp53−/−MLL-AF9 cells.25 The Trp53−/− c-Kit+ AML cells express TLR1 (supplemental Figure 9A), and similar to Trp53+/+MLL-AF9 leukemia cells, they could be forced into apoptosis (supplemental Figure 9B), and differentiation (supplemental Figure 9C), on TLR1/TLR2 activation. These findings demonstrate that activation of TLR1/TLR2 induces apoptosis and differentiation of AML cells in a p53-independent manner.
Further, to validate whether the antileukemic effects of TLR1/TLR2 activation could be extended to subgroups of AML other than the MLL-rearranged subtype, we used a murine AML1-ETO9a AML model.26 The c-Kit+ AML cells expressed both TLR1 and TLR2 (supplemental Figure 9D-E), and agonistic targeting of TLR1/TLR2 resulted in apoptosis and myeloid differentiation of the cells (supplemental Figure 9F-G). These findings demonstrate that the antileukemic effect of activating TLR1/TLR2-signaling is not confined to MLL-rearranged AML, but also extends to other AML subtypes.
TLR1/TLR2 activation mildly suppresses normal HSPCs
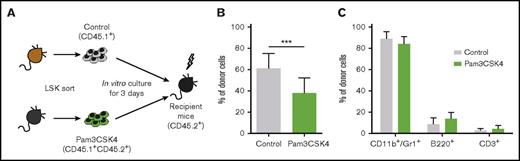
To assess the effect of activating TLR1/TLR2 on normal murine HSPCs, which, in contrast to human HSPCs, express TLR1 (supplemental Figure 10A-B), we treated Linage−Sca1+cKit+ (LSK) BM cells ex vivo with Pam3CSK4 for 3 days, which induced an increase in CD11b expression (supplemental Figure 11A). Subsequently, the cells were injected into mice in a competitive transplantation setting, using the CD45.1/CD45.2 marker system (Figure 6A). TLR1/TLR2 activation had a mild suppressive effect on long-term BM repopulating cells, evaluated 16 weeks posttransplantation (Figure 6B). No difference in lineage output in vivo was observed between the groups, demonstrating that ex vivo TLR1/TLR2 activation did not affect the lineage fate of HSPCs on BM repopulation of irradiated hosts (Figure 6C; supplemental Figure 11B).
TLR1/TLR2 activation has mild effects on long-term bone marrow repopulating cells. 10 000 sorted LSK cells were cultured for 3 days with (CD45.1+CD45.2+, Pam3CSK4 group) or without (CD45.1+, control group) Pam3CSK4. Control and Pam3CSK4-treated cells were then pooled and transplanted into lethally irradiated CD45.2+ mice along with 200 000 support cells from CD45.2+ mice (12 mice per group). (A) Schematic picture of the experiment. (B) Percentage of donor cells in bone marrow and (C) lineage distribution within donor cells in the peripheral blood 16 weeks posttransplantation. Mean values and SDs are shown. ***P < .001.
TLR1/TLR2 activation has mild effects on long-term bone marrow repopulating cells. 10 000 sorted LSK cells were cultured for 3 days with (CD45.1+CD45.2+, Pam3CSK4 group) or without (CD45.1+, control group) Pam3CSK4. Control and Pam3CSK4-treated cells were then pooled and transplanted into lethally irradiated CD45.2+ mice along with 200 000 support cells from CD45.2+ mice (12 mice per group). (A) Schematic picture of the experiment. (B) Percentage of donor cells in bone marrow and (C) lineage distribution within donor cells in the peripheral blood 16 weeks posttransplantation. Mean values and SDs are shown. ***P < .001.
TLR1/TLR2 activation in primary human AML cells results in apoptosis and differentiation
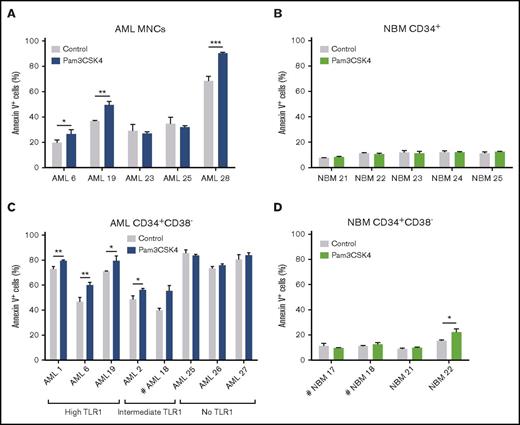
To address the effect of TLR1/TLR2 activation in AML patient cells, we next explored agonistic targeting of TLR1/TLR2 in primary human AML cells, using Pam3CSK4. TLR1/TLR2 activation of primary AML bulk MNCs resulted in increased apoptosis, whereas TLR1/TLR2 activation did not induce apoptosis of normal CD34+ BM cells (Figure 7A-B). Stimulating AML MNCs with Pam3CSK4 also induced myeloid differentiation, as evidenced by an increase in the percentage of CD15 or CD14-expressing cells (supplemental Figure 12A-B), as well as morphological changes consistent with myeloid differentiation (supplemental Figure 12C), accompanied by a weak downregulation of TLR1 (supplemental Figure 12D). Corresponding treatment of normal BM CD34+ cells revealed a milder Pam3CSK4-induced differentiation (supplemental Figure 12E-F).
TLR1/TLR2 activation results in apoptosis of primary human AML cells. AML patient samples and normal bone marrow samples were cultured for 3 days with or without (control) Pam3CSK4 and assessed for apoptosis by Annexin V+ staining. (A) Bulk mononuclear cells (MNCs) from patients with AML, (B) NBM CD34+ cells, (C) CD34+CD38− cells from patients with AML, and (D) NBM CD34+CD38− cells. High/Intermediate/No TLR1 was grouped based on TLR1 expression in AML CD34+CD38− cells (supplemental Table 1). Mean values and SDs are shown. Comparative statistical analysis based on triplicate samples was performed except for #, indicating duplicate replicates. *P < .05; **P < .01; ***P < .001.
TLR1/TLR2 activation results in apoptosis of primary human AML cells. AML patient samples and normal bone marrow samples were cultured for 3 days with or without (control) Pam3CSK4 and assessed for apoptosis by Annexin V+ staining. (A) Bulk mononuclear cells (MNCs) from patients with AML, (B) NBM CD34+ cells, (C) CD34+CD38− cells from patients with AML, and (D) NBM CD34+CD38− cells. High/Intermediate/No TLR1 was grouped based on TLR1 expression in AML CD34+CD38− cells (supplemental Table 1). Mean values and SDs are shown. Comparative statistical analysis based on triplicate samples was performed except for #, indicating duplicate replicates. *P < .05; **P < .01; ***P < .001.
Finally, we assessed the effect of TLR1/TLR2 on primitive CD34+CD38− AML cells compared with corresponding normal BM cells. In AML CD34+CD38− cells with intermediate or high TLR1 expression, we observed increased apoptosis on TLR1/TLR2 activation, whereas modest effects were observed in AML cells with no TLR1 expression and corresponding normal BM cells (Figure 7C-D). Moreover, we found more Pam3CSK4-induced differentiation of the AML CD34+CD38− cells relative to the corresponding NBM cells (supplemental Figure 12G-J).
Discussion
The TLR1/TLR2 receptor complex is a conserved sensor of the innate immune system that recognizes bacterial lipopeptides and transmits activation signals into immune cells.41 We here identified a leukemia-selective vulnerability for activation of the TLR1/TLR2 complex in murine and human AML cells. The approach to use agonistic targeting of a cell surface molecule in AML differs from other strategies that predominantly use antibodies with antagonistic and/or immune-mediated stimulatory activity.42
To date, only a few cell surface receptors that are not expressed on normal HSCs have been found to be upregulated in AML stem cell-enriched populations (eg, IL1RAP,43,44 CD25,45 CLL-1,46 and TIM-39 ). Our finding that TLR1 and TLR2 are upregulated on AML CD34+CD38− cells, but not on corresponding normal BM cells, adds TLR1 and TLR2 to this list of LSC-associated cell surface molecules in AML.
By using murine models of AML, driven by MLL-AF9 or AML1-ETO9a, along with human MA9 cells and primary AML patient cells, we found that TLR1/TLR2 activation induces apoptosis and myeloid differentiation of AML cells, whereas normal HSPCs were relatively less affected, thus highlighting the TLR1/TLR2 complex as a putative therapeutic target in AML. The finding of an enrichment of cell death pathway signatures already 3 hours after Pam3CSK4 treatment of MA9 cells suggests that induction of an apoptosis-associated transcriptional program is an early consequence of TLR1/TLR2 activation. Agonistic targeting of TLR1/TLR2 has previously been shown to sensitize acute lymphoblastic leukemia cells for vincristine-mediated cytotoxicity,47 to enhance the cytotoxic and apoptotic effects of bortezomib in myeloma cells, and to induce cell death of monocytes from tuberculosis patients,48,49 but induction of apoptosis after TLR1/TLR2 activation has not previously been described in the context of AML. The observed differentiation of AML cells is consistent with a described cellular response to TLR1/TLR2 activation in normal myeloid progenitor cells.19,50
Interestingly, although spontaneous remissions in AML are rare, an association between such events and severe bacterial infections in patients has been observed.51,52 This link has been hypothesized to depend on the activation of immune cells.53-55 Our findings suggest that other plausible mechanisms, such as the direct stimulation of TLR1/TLR2 on AML cells by bacterial lipopeptides, could have contributed to the observed remissions by causing differentiation and apoptosis of the leukemic cells.
In the search for the molecular mechanism responsible for the differentiation and apoptosis caused by TLR1/TLR2 activation, we identified increased AKT, NFκB, and p38 MAPK phosphorylation in AML cells on Pam3CSK4 stimulation. We also found that the apoptotic effect was p53-independent and that inhibition of p38 MAPK-signaling suppressed Pam3CSK4-induced Caspase 3 activation and apoptosis in AML cells, demonstrating that the apoptotic effect of Pam3CSK3 was p38 MAPK-dependent, coherent with a role of the p38 MAPK as a tumor suppressor that regulates apoptosis.56 Consistent with NFκB activation leading to differentiation of normal hematopoietic progenitor cells57 and AKT being upstream of NFκB,58,59 we confirmed that the differentiation effect of Pam3CSK4 was NFκB-dependent, using an inhibitor of NFκB-signaling. These findings suggest that TLR1/TLR2 activation drives the AML cells into a differentiation and apoptotic fate by distinct molecular mechanisms.
We also found that TLR1/TLR2 activation ex vivo suppressed the leukemia-initiating capacity of murine and human AML cells, translating into extended survival of the mice, demonstrating that agonistic targeting of TLR1/TLR2 inhibits LSCs. Although consecutive treatments of mice with Pam3CSK4 were poorly tolerated, potentially because of high inflammatory cytokine production on TLR1/TLR2 activation,60 these experiments provided proof of concept that agonistic targeting of TLR1/TLR2 has therapeutic efficacy in an in vivo AML model. Consistent with these observations, TLR1/TLR2 activation eradicated the colony formation of MA9 cells and induced differentiation and apoptosis of primary AML CD34+CD38− cells. Within normal HSPCs, ex vivo TLR1/TLR2 activation mildly suppressed the function of murine long-term BM repopulating cells, but did not result in altered lineage fate in vivo, as assessed in competitive BM repopulating assays. In line with these findings, TLR1/TLR2 activation had only mild effects on apoptosis and differentiation of human HSPCs. We speculate that the vulnerability of immature AML cells to TLR1/TLR2 activation compared with NBM cells is linked to the upregulation of TLR1 and TLR2 in the AML cells. This hypothesis was supported by a stronger response to Pam3CSK4 treatment within TLR1 expressing AML patient samples relative to samples with no TLR1 expression.
In summary, our findings reveal activation of TLR1/TLR2 signaling as a vulnerability of AML cells and provide mechanistic insights to the antileukemic effects. We demonstrate that TLR1 and TLR2 are upregulated on primitive AML patient cells and that agonistic targeting of the TLR1/TLR2 complex forces AML cells into apoptosis via p38 MAPK-signaling and differentiation via NFκB, thus highlighting TLR1 as a candidate target for therapeutic interventions in AML.
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported by the Swedish Cancer Society, the Swedish Childhood Cancer Foundation, the Crafoord Foundation, the Gunnar Nilsson Cancer Foundation, the Medical Faculty of Lund University, the Åke Wiberg Foundation, the Royal Physiographic Society of Lund, the Swedish Research Council, BioCARE, and FP7 Marie Curie.
Authorship
Contribution: M.E., P.P.-M., T.F., and M.J. designed the research; M.E., P.P.-M., R.R., M.C., C.H., G.G., and C.O.-P. performed the research; M.E., P.P.-M., and M.J. analyzed the data; V.L.L., G.J., and J.R. collected patient material and clinical data; T.V.-H., J.C., J.C.M., and B.L.E. provided new reagents; M.E. and M.J. wrote the paper; and all other authors contributed with valuable comments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marcus Järås, Department of Clinical Genetics, Klinikgatan, 28, SE-22184 Lund, Sweden; e-mail: marcus.jaras@med.lu.se.