Key Points
Core-binding factor AML can evolve from good-risk disease into aggressive disease through the gain of additional genomic aberrations.
In this unique case, an AML patient died of hypereosinophilic syndrome with solid organ infiltration of differentiated eosinophils.
Introduction
Core-binding factor acute myeloid leukemia (AML) has one of the most favorable risk profiles in AML for which allogeneic hematopoietic stem cell transplantation is not indicated as postremission therapy for patients in first complete remission (CR).1 Instead, consolidation high-dose cytarabine (HiDAC) is sufficient to achieve durable remissions in a substantial proportion of these patients.2 Here, we present a case of a patient with inv(16) AML who achieved a total of 5 CRs with various chemotherapy regimens but ultimately died of his relapsing disease, culminating in an aggressive and highly invasive leukemia with distinct eosinophilic differentiation. At each relapse, bone marrow biopsies were performed, revealing the serial acquisition of new cytogenetic abnormalities and a KRASG12D mutation. Eosinophil and AML blast cells were purified via flow cytometric cell sorting at relapses and confirmed the presence of inv(16) and the KRAS mutation in these populations. This report highlights a case in which a chemosensitive, favorable-risk AML underwent clonal evolution that was longitudinally captured and ultimately developed into unusually aggressive and lethal disease.
Case description
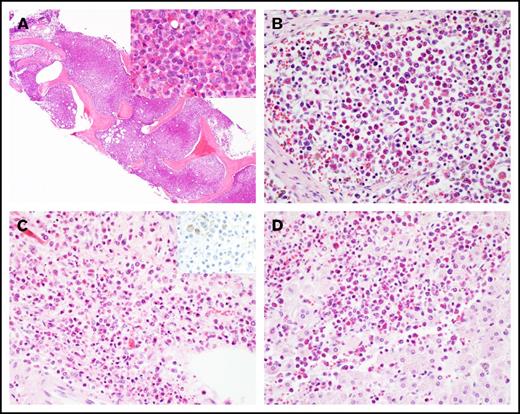
A 57-year-old man was admitted after he was found to be neutropenic during an evaluation for a right hand infection. Bone marrow biopsy revealed AML with inv(16) as the only cytogenetic abnormality (Table 1; supplemental Figure 1A). He was treated with standard induction chemotherapy with 7+3, achieved CR1, and subsequently went on to receive 2 cycles of HiDAC consolidation. His first relapse occurred ∼1 year after his initial diagnosis. He received reinduction chemotherapy with mitoxantrone, etoposide, and cytarabine and achieved CR2. A donor search was initiated, but no suitable unrelated donors were identified. His second relapse was treated with fludarabine, cytarabine, and granulocyte colony-stimulating factor (FLAG), and he achieved CR3. He suffered a third relapse almost 3 years after his initial diagnosis (supplemental Figure 1B-E), was treated with 4 cycles of decitabine, and achieved CR4 with minimal residual disease detectable by flow cytometry on his bone marrow specimen. He relapsed again ∼4 years from diagnosis (supplemental Figure 1F-I) and was rechallenged with additional cycles of decitabine but continued to exhibit a persistent rise in his circulating blast count. He received reinduction with FLAG and achieved CR5. Nearly 5 years from his initial diagnosis, the patient’s peripheral blast count began to rise, and he was found to have his fifth relapse. He was started on azacitidine and venetoclax. Two months into therapy, the patient was admitted for febrile neutropenia, dyspnea, and right upper quadrant abdominal pain and was found to have eosinophilia in the range of 2 to 7.7 K/μL. Extensive workup for reactive causes of eosinophilia was unrevealing. During his hospital course, the patient became progressively hypoxic and dyspneic. Computed tomographic imaging revealed bilateral lower lobe ground glass opacities and typhlitis. A bone marrow biopsy revealed persistent AML with marked eosinophilia. The marrow space was replaced by eosinophils and eosinophilic precursors (Figure 1A; supplemental Figure 1J-M). The patient’s clinical status continued to decline with worsening hypoxia, coagulopathy, and progression to fulminant liver failure. The patient died, and the family requested an autopsy, which concluded that the cause of death in this patient was multiorgan failure due to persistent AML with diffuse infiltration of the liver, lungs, heart, kidneys, lymph nodes, and adrenal glands with differentiated eosinophils, not AML blasts (Figure 1A-D).
Clinical course for a 57-year-old man diagnosed with inv(16) AML
| Clinical course . | Treatment . | Karyotype . | FISH, % . | NGS targeted panel . | Ratio KRAS G12D mutant: wild type . | |
|---|---|---|---|---|---|---|
| Blasts . | Eosinophils . | |||||
| Initial diagnosis | 7+3, consolidation HiDAC (2 cycles) | 46,XY, inv(16)(p13.1q22)[2/15] | 40.3 | n/d | n/d | n/d |
| First relapse | MEC | n/d | 3 | n/d | n/d | n/d |
| Second relapse | FLAG | n/d | 1.3 | n/d | 0 | 0 |
| Third relapse | Decitabine (4 cycles) | 46,XY, inv(16)(p13.1q22)[1/13] | 5 | n/d | 0 | 0 |
| Fourth relapse | FLAG | 48,XY, +8, inv(16)(p13.1q22), +20[17/20] | 42-45 | No mutations called | 0.02 | QNS |
| Fifth relapse | Azacitidine + venetoclax | n/d | 47 | n/d | 0.844 | QNS |
| Disease progression | None | 46,XY, add(9)(p24), inv(16)(p13.1q22), t(17;22)(q23;q13)[20] | 48 | KRAS G12D | 0.912 | 0.877 |
| Clinical course . | Treatment . | Karyotype . | FISH, % . | NGS targeted panel . | Ratio KRAS G12D mutant: wild type . | |
|---|---|---|---|---|---|---|
| Blasts . | Eosinophils . | |||||
| Initial diagnosis | 7+3, consolidation HiDAC (2 cycles) | 46,XY, inv(16)(p13.1q22)[2/15] | 40.3 | n/d | n/d | n/d |
| First relapse | MEC | n/d | 3 | n/d | n/d | n/d |
| Second relapse | FLAG | n/d | 1.3 | n/d | 0 | 0 |
| Third relapse | Decitabine (4 cycles) | 46,XY, inv(16)(p13.1q22)[1/13] | 5 | n/d | 0 | 0 |
| Fourth relapse | FLAG | 48,XY, +8, inv(16)(p13.1q22), +20[17/20] | 42-45 | No mutations called | 0.02 | QNS |
| Fifth relapse | Azacitidine + venetoclax | n/d | 47 | n/d | 0.844 | QNS |
| Disease progression | None | 46,XY, add(9)(p24), inv(16)(p13.1q22), t(17;22)(q23;q13)[20] | 48 | KRAS G12D | 0.912 | 0.877 |
At each relapse, treatment, karyotype, fluorescence in situ hybridization (FISH), mutations detected by NGS targeted panel, and ddPCR KRAS mutation detection are indicated when available.
MEC, mitoxantrone, etoposide, and cytarabine; n/d, not done; QNS, quantity not sufficient.
Histology of hypereosinophilic syndrome in AML. (A) Marked eosinophilia in the bone marrow at the last relapse (original magnification ×40; inset ×400; hematoxylin and eosin stain). Eosinophilic infiltrate in (B) lymph node, (C) adrenal medulla (inset: CD34 immunostain), and (D) liver at autopsy. Original magnification by ×400, hematoxylin and eosin stain, except as noted.
Histology of hypereosinophilic syndrome in AML. (A) Marked eosinophilia in the bone marrow at the last relapse (original magnification ×40; inset ×400; hematoxylin and eosin stain). Eosinophilic infiltrate in (B) lymph node, (C) adrenal medulla (inset: CD34 immunostain), and (D) liver at autopsy. Original magnification by ×400, hematoxylin and eosin stain, except as noted.
Methods
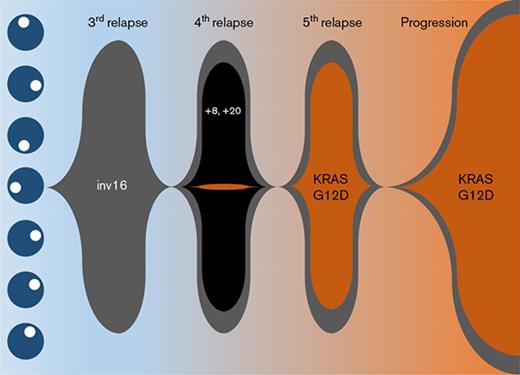
Cytogenetic characterization of the AML at diagnosis was notable only for inv(16) (Table 1), which was confirmed by FISH analysis. This abnormality persisted throughout the entire clinical course. At fourth relapse, clonal evolution with gain of chromosomes 8 and 20 was observed (supplemental Figure 2A). Accordingly, there was an immunophenotypic switch from partial CD19 positivity to CD56 positivity on the blasts (supplemental Figure 1D,I). However, these trisomies and immunophenotypic changes were not retained in the final relapse sample (supplemental Figure 1M). Instead, there were new chromosomal abnormalities detected with 46,XY,add(9)(p24),t(17;22)(q23;q13) in addition to inv(16)(p13.1q22) in 48% of cells analyzed (supplemental Figure 2B).
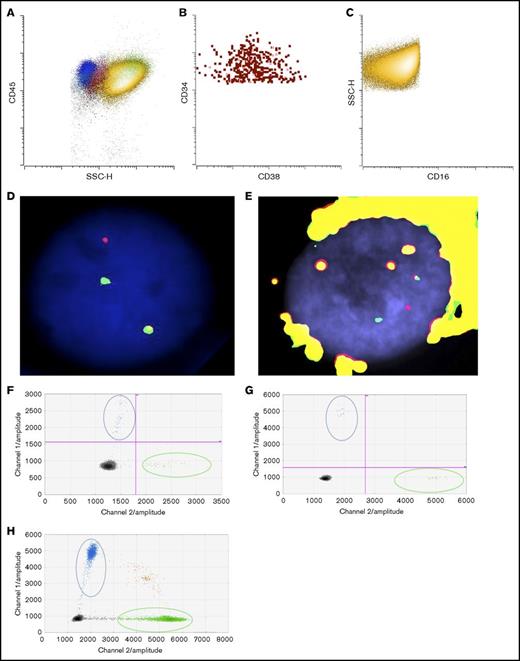
To determine if the patient’s eosinophils were derived from the leukemic clone, we flow-sorted eosinophils from the bone marrow at the time of final disease progression to examine if they also harbored inv(16). As shown in Figure 2A-C, there was a predominant CD16−CD45bright eosinophil population (in yellow) as well as a small but discrete CD34+ blast population (in red). We successfully obtained separate blast (Figure 2B) and eosinophil (Figure 2C) populations with high purity (>99%). FISH studies confirmed inv(16) in both blast (Figure 2D) and eosinophil populations (Figure 2E).
Detection of KRASG12Dmutation and inv(16) in sorted blasts and eosinophils. (A) Dot plot of bone marrow cells (yellow: eosinophils; red: blasts; blue: lymphocytes; pink: monocytes; green: neutrophils). (B) Sorted blasts (CD45dimCD34+CD15−CD117+SSClo). (C) Sorted eosinophils (CD45+CD34−CD117−CD15+SSChi). FISH studies using CBFB break-apart probes (Abbott Molecular, Des Plaines, IL) showing a split signal pattern of CBFB probes (ie, inv(16)), in (D) sorted blasts and in (E) eosinophils. Detection of KRASG12D in (F) sorted blasts, (G) eosinophils, and (H) FFPE adrenal tissue by ddPCR. Blue dots represent mutant droplets; green dots represent wild-type droplets, and red or gray dots represent empty droplets. SSC-H, side-scatter height.
Detection of KRASG12Dmutation and inv(16) in sorted blasts and eosinophils. (A) Dot plot of bone marrow cells (yellow: eosinophils; red: blasts; blue: lymphocytes; pink: monocytes; green: neutrophils). (B) Sorted blasts (CD45dimCD34+CD15−CD117+SSClo). (C) Sorted eosinophils (CD45+CD34−CD117−CD15+SSChi). FISH studies using CBFB break-apart probes (Abbott Molecular, Des Plaines, IL) showing a split signal pattern of CBFB probes (ie, inv(16)), in (D) sorted blasts and in (E) eosinophils. Detection of KRASG12D in (F) sorted blasts, (G) eosinophils, and (H) FFPE adrenal tissue by ddPCR. Blue dots represent mutant droplets; green dots represent wild-type droplets, and red or gray dots represent empty droplets. SSC-H, side-scatter height.
Targeted sequencing studies using a 49-gene hematologic malignancy–focused next-generation sequencing (NGS) panel revealed a KRASG12D mutation at final disease progression, which was not called at the patient’s fourth relapse (Table 1). To investigate whether the KRAS mutation was present at low levels during his prior relapses, we obtained viably cryopreserved bone marrow aspirate specimens collected during his prior relapses from our institutional, institutional review board–approved biospecimen bank. We successfully purified the patient’s blast and eosinophil populations from the samples at various time points. Eosinophil yields at the patient’s fourth and fifth relapses were poor likely due to the detrimental impact of cryopreservation on eosinophils. Nevertheless, droplet digital polymerase chain reaction (ddPCR) was able to detect the presence of the KRASG12D mutation as early as the fourth relapse in the purified blasts albeit at very low levels, when our targeted sequencing panel did not identify any mutations. At the time of the fifth relapse, the representation of this mutant clone expanded dramatically, such that the ratio of mutant droplets to wild-type droplets was >0.8, and this continued to rise at the time of his final disease progression, where it was nearly 0.9 in both blasts and eosinophils (Table 1; Figure 2F-G).
In addition to detection of the KRASG12D mutation in bone marrow–derived eosinophils, we sought to confirm that the eosinophilic solid organ infiltration was also derived from the leukemic clone. Genomic DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) adrenal gland tissue processed at the time of autopsy. CD34 and CD117 immunostaining of adrenal tissue confirmed that the infiltrate was devoid of AML blasts but primarily composed of mature eosinophils (Figure 1C inset and data not shown). Similar to flow-sorted blast and eosinophil populations derived from the bone marrow, ddPCR detected the KRASG12D mutation in the adrenal gland, with a mutant-to-wild-type ratio of 0.078. By extrapolation, ∼15% of the total cells harbor this mutation in the adrenal tissue. We attempted FISH studies on the adrenal gland tissue, but artifacts owing to the highly refractile nature of the eosinophilic granules precluded definitive interpretation of these studies. Furthermore, we also attempted to measure CBFB/MYH11 RNA transcripts generated by the inv(16) translocation, but insufficient high-quality RNA was extracted from FFPE adrenal tissue. Nevertheless, robust detection of the KRASG12D mutation by ddPCR and the relative absence of AML blasts by immunostaining of the adrenal tissue suggest that the solid organ eosinophilic infiltrate ultimately was leukemic in origin.
Results and discussion
We present a unique case of a patient diagnosed with inv(16) AML. Over the 5-year course of this patient’s AML treatment from initial diagnosis until his death, we were able to track the clonal evolution of his disease (supplemental Figure 2). Although allogeneic hematopoietic stem cell transplantation was not feasible due to a lack of donor options for this patient, he was remarkably able to achieve 5 CRs with multiple chemotherapy regimens. This illustrates how chemosensitive core-binding factor AMLs can be. However, despite this chemosensitivity, prolonged persistence of the inv(16)-driven founding clone in this patient allowed sufficient time for the acquisition of additional cytogenetic abnormalities, and in particular, the acquisition of a new KRASG12D-mutant clone that appeared to gain clonal dominance under the selection pressure of chemotherapy and drive eosinophilic differentiation, resulting in the extensive solid-organ eosinophilic infiltration that caused this patient’s multisystem organ failure and ultimately his death.
Core-binding factor AML is known to be associated with extramedullary involvement. In 1 of the largest studies of extramedullary disease, Ganzel et al reported that, out of 200 patients with favorable risk AML defined as having inv(16)/t(16;16)/del(16q) or t(8;21), 13% of these patients had extramedullary disease.3 Other case reports have demonstrated the presence of the inv(16) cytogenetic abnormality in bone marrow–derived eosinophils from AML patients,4,5 but our report is the first to show that the solid-organ eosinophilic infiltrate also originates from an evolved leukemic clone through ddPCR-based detection of a KRASG12D mutation in adrenal tissue. Additional studies are also needed to assess the relative frequency with which solid organ eosinophilic infiltration occurs in AML.
The expansion of this patient’s KRAS-mutant clone is also noteworthy. RAS mutations occur at a frequency of ∼10% to 15% in AML, with the majority of the mutations arising in NRAS rather than KRAS.6,7 Preclinical mouse models of RAS-mutant AML have demonstrated that KRASG12D mutations are more aggressive and lethal than NRASG12D mutations.8 Although these RAS mutations have not yet demonstrated independent prognostic significance, patients with RAS-mutant AML may have a better response to HiDAC as postremission therapy.9,10
In addition, the application of multiparametric flow cytometry to purify discrete AML blast and eosinophil populations coupled with ddPCR proved to be instrumental in (1) determining the etiology of this patient’s hypereosinophilia, (2) capturing the subclonal KRASG12D mutation at a point in time when NGS panels were unable to detect the mutation, and (3) deconvoluting the clonal evolution and clonal architecture of this particular patient’s AML. In conclusion, this unique case illustrates how a highly chemosensitive AML can undergo clonal evolution that can be followed longitudinally over a patient’s clinical course from diagnosis through progression into a lethal disease with eosinophilic differentiation.
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported by a Young Investigator Award from the Conquer Cancer Foundation (S.F.C.), a Career Development Program Fellow Award from the Leukemia and Lymphoma Society (S.F.C.), and a startup fund from the Department of Pathology at the Memorial Sloan Kettering Cancer Center (W.X.). This research was also funded in part through the National Institutes of Health, National Cancer Institute Cancer Center Support grant P30 CA008748.
Authorship
Contribution: S.F.C., W.X., M.Y., M.O., R.J.D., and J.J.P. wrote the manuscript; J.B. coordinated sequencing and immunohistochemistry studies; M.Y., W.X., P.K., M.R., Y.Z., and M.S.T. performed diagnostic tests and analyzed the data; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sheng F. Cai, Leukemia Service, Department of Medicine, Memorial Sloan Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: cais1@mskcc.org.
References
Author notes
W.X. and M.Y. contributed equally to this work.