Key Points
The 3 established HbF genetic loci can be summarized into 1 quantitative variable, g(HbF), in SCD and influence markers of SCD severity.
g(HbF) provides a quantitative marker for the genetic component of HbF% variability, potentially useful in genetic and clinical studies in SCD.
Abstract
Fetal hemoglobin (HbF) is a strong modifier of sickle cell disease (SCD) severity and is associated with 3 common genetic loci. Quantifying the genetic effects of the 3 loci would specifically address the benefits of HbF increases in patients. Here, we have applied statistical methods using the most representative variants: rs1427407 and rs6545816 in BCL11A, rs66650371 (3-bp deletion) and rs9376090 in HMIP-2A, rs9494142 and rs9494145 in HMIP-2B, and rs7482144 (Xmn1-HBG2 in the β-globin locus) to create g(HbF), a genetic quantitative variable for HbF in SCD. Only patients aged ≥5 years with complete genotype and HbF data were studied. Five hundred eighty-one patients with hemoglobin SS (HbSS) or HbSβ0 thalassemia formed the “discovery” cohort. Multiple linear regression modeling rationalized the 7 variants down to 4 markers (rs6545816, rs1427407, rs66650371, and rs7482144) each independently contributing HbF-boosting alleles, together accounting for 21.8% of HbF variability (r2) in the HbSS or HbSβ0 patients. The model was replicated with consistent r2 in 2 different cohorts: 27.5% in HbSC patients (N = 186) and 23% in 994 Tanzanian HbSS patients. g(HbF), our 4-variant model, provides a robust approach to account for the genetic component of HbF in SCD and is of potential utility in sickle genetic and clinical studies.
Introduction
High fetal hemoglobin (HbF) levels are clinically beneficial in sickle cell disease (SCD), being associated with longer survival1 and lower pain rates.2 Patients with SCD have higher HbF levels compared with nonaffected adults and, within SCD, HbF levels are higher in hemoglobin SS (HbSS) compared with HbSC individuals.3 One component of HbF variability relates to the expanded erythron secondary to chronic hemolysis, and preferential survival of HbF-containing red cell precursors (F cells).4,5 A second component is the innate ability for HbF synthesis based on genetic variants at 3 quantitative trait loci: BCL11A on chromosome 2p, HMIP-2 on chromosome 6q, and Xmn1-HBG2 (rs7482144) on chromosome 11p. Dependent upon the genetic variants investigated and analysis performed, such variants were found to account for between 8 and ∼20% of the HbF variability in SCD in studies from the United Kingdom, United States, Brazil, Tanzania, and Cameroon.6-13 This genetic component is likely to account for much of the variability in HbF levels in SCD patients. Consequently, it may be helpful to quantify and summarize the effects of the respective genetic loci into a single genetic variable to capture the essence of genetic disease alleviation through the HbF mechanism. Here, we present such a genetic HbF summary variable, g(HbF), which will be a useful parameter to use as a covariate in genetic, biological, and clinical studies in diverse SCD populations.
Subjects and methods
British patients are part of the South East London sickle gene bank (King’s College Hospital, Guys and St Thomas’ Hospitals Trust, Lewisham Hospital, and Queen Elizabeth Hospital Woolwich). Written informed consent was obtained through 3 approved study protocols (LREC 01-083, 07/H0606/165, and 12/LO/1610) and research conducted in accordance with the Helsinki Declaration (1975, as revised 2008).
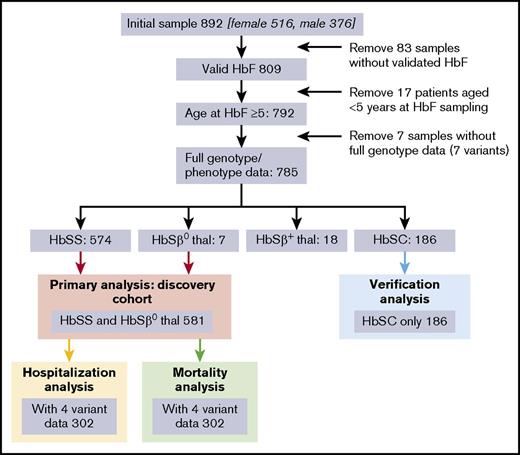
Eight hundred ninety-two patients consented to the study, of which 785 aged over 5 years had a full dataset (genotypes and phenotype). These 785 comprised 581 with HbSS or HbSβ0 (the “discovery cohort” used for the primary analysis), 186 HbSC (the “validation” cohort), and 18 HbSβ+ thalassemia (Figure 1). Additional validation was performed in the Muhimbili HbSS cohort, Tanzania (N = 994).10
Genetic variants and genotyping
We assembled an initial set of 7 known (and widely replicated) HbF modifier variants, prioritizing those where additional functional evidence had been generated (Table 1): BCL11A-rs142740714 and rs654581610 ; HMIP-2A-rs6665037115,16 and rs937609017 ; HMIP-2B-rs949414517,18 and rs949414216 ; Xmn1-HBG2-rs7482144.6,19,20
Seven variants representing the 3 major HbF quantitative trait loci that were rationalized to a 4-variant model (g(HbF)) to represent the genetic component of HbF in SCD
| Gene . | Variant . | HbF-boosting allele . | Allele frequency . | g(HbF) model . | ||
|---|---|---|---|---|---|---|
| London, United Kingdom . | Dar es Salaam, Tanzania . | Coefficient (β) (95% confidence interval) . | P . | |||
| BCL11A (Chr 2) | rs6545816 (A>C) | C | 0.34 | 0.36 | 0.14 (0.08-0.19) | <.001 |
| rs1427407 (G>T) | T | 0.26 | 0.21 | 0.30 (0.26-0.35) | <.001 | |
| HMIP (HBSL1-MYB intergenic polymorphism on Chr 6) | ||||||
| 2A | rs9376090 (T>C) | C | 0.01 | |||
| rs66650371 (TAC>—) | 3bp del | 0.04 | 0.02 | 0.13 (0.08-0.17) | <.001 | |
| 2B | rs9494142 (T>C) | C | 0.05 | |||
| rs9494145 (T>C) | C | 0.05 | ||||
| HBG2-Xmn1 (β-globin gene cluster on Chr 11) | rs7482144 (G>A) | A | 0.07 | 0.01 | 0.10 (0.05-0.15) | <.001 |
| Gene . | Variant . | HbF-boosting allele . | Allele frequency . | g(HbF) model . | ||
|---|---|---|---|---|---|---|
| London, United Kingdom . | Dar es Salaam, Tanzania . | Coefficient (β) (95% confidence interval) . | P . | |||
| BCL11A (Chr 2) | rs6545816 (A>C) | C | 0.34 | 0.36 | 0.14 (0.08-0.19) | <.001 |
| rs1427407 (G>T) | T | 0.26 | 0.21 | 0.30 (0.26-0.35) | <.001 | |
| HMIP (HBSL1-MYB intergenic polymorphism on Chr 6) | ||||||
| 2A | rs9376090 (T>C) | C | 0.01 | |||
| rs66650371 (TAC>—) | 3bp del | 0.04 | 0.02 | 0.13 (0.08-0.17) | <.001 | |
| 2B | rs9494142 (T>C) | C | 0.05 | |||
| rs9494145 (T>C) | C | 0.05 | ||||
| HBG2-Xmn1 (β-globin gene cluster on Chr 11) | rs7482144 (G>A) | A | 0.07 | 0.01 | 0.10 (0.05-0.15) | <.001 |
A combination of 3 genotyping methodologies was used: (1) “manual” genotyping in the laboratory (all variants) by the TaqMan procedure, except rs66650371, which was assayed by capillary electrophoresis (Applied Biosystems, Foster City, CA), as previously described17 ; (2) a genome-wide chip (Illumina Infinium Multi-Ethnic Genotyping Array); (3) imputation with public and in-house reference haplotypes (see supplemental Methods).
Phenotypes
HbF% (measured by high-performance liquid chromatography; BioRad Variant II), no red cell transfusion for >3 months, off hydroxyurea for >3 months, and not pregnant, were retrospectively collected. For the 581 HbSS/HbSβ0 discovery set, median HbF was 4.5% (interquartile range: 1.9% to 8.8%) (supplemental Figure 1).
We estimated global disease severity using “hospitalization rate” as a measure of pain frequency, mortality, and laboratory results. Mean hospitalization rates were calculated for King’s College Hospital adults over 10 years (2004-2013), dividing an individual’s number of hematology admissions by the number of observed years. For the 302 patients with HbSS/HbSβ0, median mean hospitalization rate was 0.25 per year (interquartile range: 0-0.71) (supplemental Figure 2). Mortality outcome was available for the 302 adults (1 January 2004 to 31 July 2015). Steady-state laboratory values (hemoglobin, white blood cells) over a 10-year period (2004-2013) were averaged for 278 patients.
Building and validating the genetic model for HbF%
Genetic association between the 7 genetic variants (as normalized genotype scores) and HbF [ln(%HbF)] was investigated by linear regression (using STATA12) under an additive allelic model.
Manual linear regression modeling was carried out in the HbSS/HbSβ0 thalassemia “discovery group” (see supplemental Methods). We then validated the model, g(HbF), in 2 replication groups: (1) our own HbSC subgroup (N = 186) and (2) a Tanzanian HbSS cohort (N = 994).10
Testing for association of g(HbF) with clinical severity
Tests for genetic association of individual DNA variants with Ln(HbF%) were performed by linear regression (in STATA) with age (at sampling) and sex as covariates in an additive allelic model. Age was squared, as this was better correlated with outcome. See supplemental Methods.
Results
Summary variables combining genotypes across HbF modifier loci have been found to be associated with clinical severity in β-thalassemia21 and have also been explored in SCD.10,22-24 To represent the relationship between genetic factors and HbF more accurately and to build a summary variable that is robust across diverse SCD cohorts, we used regression modeling of the effect of 7 known modifier variants (Table 1) on HbF levels in 581 SCD patients with HbSS and HbSβ0 genotypes. We targeted genetic variants at the 3 major HbF loci that have been widely replicated and implicated as causative genetic variants. Preliminary analysis using basic regression with age/sex only yielded a model with r2 = 0.11. Adding α-thalassemia (in a subset of patients, N = 273, with α-globin status available) showed that α-thalassemia was not associated with HbF levels in our cohort (r2 = 0.11 with α-globin status, P = .23 for α-globin status). We therefore did not pursue using α-globin status. Basic regression with the 7 genetic variants only produced a model with r2 = 0.23. Putting age, sex, and the 7 genetic variants together in the model increased the r2 to 0.3167 (supplemental Figure 3). As age and sex are roughly orthogonal to the variants, our subsequent analyses did not control for age/sex.
Final regression analysis resulted in a model utilizing 4 variants: rs1427407, rs6545816 (both BCL11A), rs66650371 (HMIP-2A), and rs7482144 (Xmn1-HBG2) (Table 1). rs9376090, rs9494142, or rs9494145 (all at HMIP-2) did not improve the model and were considered redundant. Applying this model, the predicted Ln(HbF%), g(HbF), would be calculated as g(HbF) = 1.89 + 0.14 × rs6545816 + 0.3 × rs1427407 + 0.13 × rs66650371 + 0.1 × rs7482144 (genotype for each variant =0, 1, or 2, according to the number of HbF-boosting alleles).
To calculate a genetically predicted HbF% (rather than Ln(HbF%)), the reader should antilog the g(HbF) formula. Nevertheless, the formula stated above should be used for generating the covariate for statistical analyses.
g(HbF) underlies 22% (r2= 0.2178, P < .0001) of the variability in HbF levels in our discovery group, and confirming its robustness, 23% in the Muhimbili “replication group” (N = 994) and 27.5% in HbSC patients (Table 1). In HbSC disease, the comparatively large effect of g(HbF) is likely due to the less severe pathology and thus smaller influence of nongenetic factors.
HbF levels affect the severity of SCD; patients with higher levels of HbF have fewer complications and live longer.1,2 We tested the influence of g(HbF) on hospitalization rate in HbSS/HbSβ0 patients and detected tentative association (N = 304, β = 0.47, P = .031), suggesting that a 2.7-fold increase in g(HbF) would result in a 38% decrease in hospitalization frequency. Nevertheless, the g(HbF) for frequently admitted patients was not significantly changed. g(HbF) was, however, associated with hemoglobin (N = 278, β = 17.871, P < .001). We found no association of g(HbF) with mortality or white blood cells.
Discussion
Our cohort has potential power to investigate the influence of g(HbF) on global measures of disease severity. International collaboration, larger sample sizes, adding new loci as they are discovered, and development of the formula will be required to realize the utility of the g(HbF) variable. We saw no significant benefit for including the HMIP-2B locus17 in g(HbF). This will be revisited once the underlying functional variant has been identified.
We believe that estimating g(HbF), or similar genetic summary variables, will add significant value to genetic and clinical studies, either to test the influence of genetic modifiers on outcomes or to act as a covariate to adjust for such effects. The comparatively larger value of g(HbF) for HbSC patients suggests that it is able to isolate the genetic component of HbF% from the component reactive to disease severity. Using a preset formula, such as the one proposed here, will be especially useful in smaller and medium-size cohorts or clinical trials, where de novo modeling is meaningless.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Clive Stringer (System Delivery Manager, King’s College Hospital) for help in data extraction from electronic patient records. They also thank Charles Curtis and Sanghyuck Lee for their work processing the samples for the Illumina MEGA chip.
This work was supported by the Medical Research Council, United Kingdom G0001249 and ID62593 (S.L.T.) and a grant from Shire Pharmaceuticals (S.M. and S.L.T.). This work is also supported by the University College London Hospitals Biomedical Research Centre, and by awards establishing the Farr Institute of Health Informatics Research at UCL Partners, from the Medical Research Council, Arthritis Research UK, British Heart Foundation, Cancer Research UK, Chief Scientist Office, Economic and Social Research Council, Engineering and Physical Sciences Research Council, National Institute for Health Research, National Institute for Social Care and Health Research, and Wellcome Trust grant MR/K006584/1 (S. Newhouse).
Authorship
Contribution: S.M., T.F., and S.L.T. designed the research study; K.G., H.R., J.H., and S.L.T. collected data; K.G., H.R., N.A., and N.S. performed experiments; T.F., K.G., S.M., H.P., and S. Newhouse analyzed the data; K.G., S.M., and S.L.T. wrote the paper; S. Nkya, J. Makani, R.Z.S., and J. Mgaya provided data from the Muhimbili Sickle Cell Biorepository (Dar es Salaam, Tanzania) for analysis; M. Allman, R.K.-A., D.C.R., S.S.-S., T.Y., and M. Awogbade provided clinical materials and data; and all authors participated in editing the final version of paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.L.T. is Sickle Cell Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD.
Correspondence: Swee Lay Thein, Sickle Cell Branch, National Heart, Lung, and Blood Institute, The National Institutes of Health, Building 10-CRC, Room 6S241, 10 Center Dr, Bethesda, MD 20892; e-mail: sl.thein@nih.gov; and Kate Gardner, Red Cell Biology Programme, King’s College London, Rayne Institute, 123 Coldharbour Ln, London SE5 9NU, United Kingdom; e-mail: kate.gardner@doctors.org.uk.
References
Author notes
S.M. and S.L.T. are joint senior authors.