Abstract
HLA haploidentical hematopoietic cell transplantation (haplo-HCT) using posttransplantation cyclophosphamide (PT-Cy) is an alternative strategy when a matched sibling donor (MSD) is not available. We performed a systematic review and meta-analysis to compare the outcomes of MSD vs haplo-HCT. Eleven studies (1410 haplo-HCT and 6396 MSD recipients) were meta-analyzed. All studies were retrospective and high quality, and 9 were multicenter. Haplo-HCT was associated with ~50% lower risk of chronic graft-versus-host disease (GVHD) (hazard ratio [HR], 0.55; 95% confidence interval [CI], 0.41-0.74), but higher risk of nonrelapse mortality (HR, 1.36; 95% CI, 1.12-1.66). Relapse, survival, acute GVHD, and GVHD-free relapse-free survival were not significantly different between the groups. Deciphering the relative contribution of PT-Cy and HLA disparity to the observed outcome differences between the groups requires further research.
Introduction
Although an HLA-matched sibling is considered the optimal donor in allogeneic hematopoietic cell transplantation (HCT),1 haploidentical HCT (haplo-HCT) has emerged as a major alternative strategy.2,3 However, a consensus opinion regarding the outcomes of HCT using a matched sibling donor (MSD) vs a haploidentical donor has not been achieved, due in part to the heterogeneity of previous studies and the lack of a randomized trial. To address this knowledge gap, we performed a systematic review and meta-analysis and generated the highest level of evidence using the available data.
Methods
PubMed and Scopus were searched through 1 June 2019 using the following MeSH terms and keywords: “graft-versus-host disease” (GVHD), “haploidentical transplant,” “matched related donor,” and “matched sibling donor.” The identified articles were screened for full-text review based on title and abstract. The bibliography of the eligible articles was reviewed to identify additional studies. The included studies had to be in English, published as peer-reviewed articles, used posttransplant cyclophosphamide (PT-Cy) in haplo-HCT recipients, and reported ≥1 of the following outcomes in both MSD and haplo-HCT groups: acute GVHD, chronic GVHD (cGVHD), disease-free survival (DFS), overall survival (OS), nonrelapse mortality (NRM), relapse, and GVHD-free relapse-free survival (GRFS). Data were independently extracted by 2 authors, and disagreements were resolved through discussion with the last author. Data synthesis was performed according to the guidelines for systematic reviews and meta-analyses.4 We chose a random-effects model (R version 3.6.0) because of the expected heterogeneity of studies in their populations, exposures, and interventions.5
Between-study heterogeneity was quantified by the I2 statistic, which estimates the percentage of total variation across studies that is due to heterogeneity rather than chance.6 Funnel plots and Egger’s regression were used to assess publication bias,7 which arises when studies with statistically significant results are more likely to be published than “negative” studies. Publication bias is more likely to affect smaller studies and can be assessed by Egger’s test for the asymmetry of the scatterplot of treatment effect against sample size (funnel plot). We conducted a sensitivity analysis by excluding potential outliers8 and then comparing the pooled effect size with vs without outliers. A study was considered an outlier if its standardized residual was >3 in absolute magnitude.9 The standardized residual for a given study is defined as the difference between that study’s effect size and the combined effect size divided by the standard deviation of effect sizes across all studies. Considering an expected normal distribution for standardized residuals around the combined effect size, studies that fall near the tails of the histogram are considered outliers.
Two authors independently assessed the risk of bias in the included studies using the Newcastle-Ottawa checklist,10 evaluating internal validity threats in 3 domains: selection bias, misclassification, and noncomparability. Points were allocated based on the selection process of cohorts (0-4), comparability of cohorts (0-2), and the assessment of outcomes of study participants (0-3). We considered total scores of 0 to 3, 4 to 6, and 7 to 9 as low, moderate, and high quality, respectively.
Results and discussion
A total of 20 articles was identified for the full-text review, 11 of which were selected for final analysis (1410 haplo-HCT and 6396 MSD recipients)11-21 (Table 1; supplemental Figure 1). One study15 reported outcomes for patients aged <55 years and ≥55 years separately and was considered 2 studies. We used as much information as possible from each study. For example, if studies A and B had overlapping populations but study A reported an outcome that study B did not, we kept both studies but used A for its unique outcome and B for the other outcomes.
Study characteristics
| Study (year) . | n . | Age, median (range), y . | Male, % . | Disease . | Conditioning, RI/MA, % . | Graft source, BM/PB, % . | GVHD prophylaxis . | F/U, median (range), mo . |
|---|---|---|---|---|---|---|---|---|
| Ahmed et al (2019)20,*,† | ||||||||
| Haplo | 139 | 33 (19-69) | 58 | HL | 100/0 | 70/30 | PT-Cy based | 52 (2-101) |
| MSD | 457 | 33 (18-66) | 56 | HL | 100/0 | 4/96 | CNI + (MMF 31%, MTX 48%, other 21%) | 37 (5-109) |
| Bashey et al (2016)21 | 45 (13-120) | |||||||
| Haplo | 116 | 51 (20-74) | 54 | Any | 60/40 | 55/45 | PT-Cy + Tac/MMF | |
| MSD | 181 | 53 (19-74) | 58 | Any | 46/54 | 1/99 | Tac/MTX | |
| Burroughs et al (2008)11,* | ||||||||
| Haplo | 28 | 32 (14-62) | 46 | HL | 100/0 | 100/0 | PT-Cy + Tac/MMF | 22 (4-62) |
| MSD | 38 | 33 (17-64) | 53 | HL | 100/0 | 0/100 | MMF/CNI | 24 (11-87) |
| Devillier et al (2018)17 | 26 (4-77) | |||||||
| Haplo | 33 | ≥60 | NR | AML | 100/0 | 0/100 | PT-Cy based | |
| MSD | 31 | ≥60 | NR | AML | 84/16 | 6/94 | NR | |
| Dietrich et al (2016)19,*,† | NR | |||||||
| Haplo | 59 | ≥18 | NR | NHL | 78/22 | NR | PT-Cy + (MMF/CNI 86%, CNI 5%, NR 9%) | |
| MSD | 2024 | ≥18 | NR | NHL | NR | NR | NR | |
| Dreger et al (2019)16,*,† | ||||||||
| Haplo | 132 | 58 (20-75) | 65 | DLBCL | 100/0 | 75/25 | PT-Cy ± CNI/MMF | 49 (12-73) |
| MSD | 525 | 55 (19-73) | 62 | DLBCL | 100/0 | 2/98 | CNI + (MMF 36%, MTX 45%, other 19%) | 48 (2-97) |
| Gauthier et al (2018)18,*,† | ||||||||
| Haplo | 61 | 29 (17-68) | 54 | HL | 100/0 | 51/49 | PT-Cy + CNI/MMF | 24 (3-58) |
| MSD | 90 | 32 (12-67) | 63 | HL | 100/0 | 14/86 | CNI + (none 39%, MMF 35%, MTX 25%) | 24 (3-70) |
| Ghosh et al (2016)14,*,† | ||||||||
| Haplo | 180 | 55 (18-75) | 64 | HL, NHL | 100/0 | 93/7 | PT-Cy ± CNI/MMF | 37 (6-73) |
| MSD | 807 | 54 (18-77) | 61 | HL, NHL | 100/0 | 2/98 | CNI + (MMF 31%, MTX 55%, other 14%) | 36 (3-76) |
| Martinez (2017)12 *,† | ||||||||
| Haplo | 98 | 31 (18-68) | 57 | HL | 90/10 | 61/39 | PT-Cy + CNI + (MMF 94%, other 6%) | 27 (1-64) |
| MSD | 338 | 32 (18-67) | 43 | HL | 69/31 | 10/89 | CNI + (MMF 27%, MTX 39%, other 34%) | 27 (1-76) |
| Robinson et al (2018)15,*,†,‡ | NR | |||||||
| Haplo | 218 | 41 (18-55) | 59 | AML, ALL | 40/60 | 57/43 | PT-Cy + CNI/MMF | |
| MSD | 843 | 42 (18-55) | 52 | AML, ALL | 16/84 | 12/88 | CNI + (MMF 20%, MTX 64%, none 16%) | |
| Robinson et al (2018)15,*,†,§ | NR | |||||||
| Haplo | 218 | 63 (55-76) | 59 | AML, ALL | 72/28 | 70/30 | PT-Cy + CNI/MMF | |
| MSD | 864 | 63 (55-76) | 58 | AML, ALL | 73/27 | 6/94 | CNI + (MMF 38%, MTX 45%, none 18%) | |
| Solh et al (2016)13 | 46 (12-123) | |||||||
| Haplo | 128 | 50 (19-74) | 54 | Any | 59/41 | 52/48 | PT-Cy + Tac/MMF | |
| MSD | 198 | 53 (18-77) | 59 | Any | 47/53 | 2/98 | Tac/MTX 66%, Tac/MMF 25%, other 9% |
| Study (year) . | n . | Age, median (range), y . | Male, % . | Disease . | Conditioning, RI/MA, % . | Graft source, BM/PB, % . | GVHD prophylaxis . | F/U, median (range), mo . |
|---|---|---|---|---|---|---|---|---|
| Ahmed et al (2019)20,*,† | ||||||||
| Haplo | 139 | 33 (19-69) | 58 | HL | 100/0 | 70/30 | PT-Cy based | 52 (2-101) |
| MSD | 457 | 33 (18-66) | 56 | HL | 100/0 | 4/96 | CNI + (MMF 31%, MTX 48%, other 21%) | 37 (5-109) |
| Bashey et al (2016)21 | 45 (13-120) | |||||||
| Haplo | 116 | 51 (20-74) | 54 | Any | 60/40 | 55/45 | PT-Cy + Tac/MMF | |
| MSD | 181 | 53 (19-74) | 58 | Any | 46/54 | 1/99 | Tac/MTX | |
| Burroughs et al (2008)11,* | ||||||||
| Haplo | 28 | 32 (14-62) | 46 | HL | 100/0 | 100/0 | PT-Cy + Tac/MMF | 22 (4-62) |
| MSD | 38 | 33 (17-64) | 53 | HL | 100/0 | 0/100 | MMF/CNI | 24 (11-87) |
| Devillier et al (2018)17 | 26 (4-77) | |||||||
| Haplo | 33 | ≥60 | NR | AML | 100/0 | 0/100 | PT-Cy based | |
| MSD | 31 | ≥60 | NR | AML | 84/16 | 6/94 | NR | |
| Dietrich et al (2016)19,*,† | NR | |||||||
| Haplo | 59 | ≥18 | NR | NHL | 78/22 | NR | PT-Cy + (MMF/CNI 86%, CNI 5%, NR 9%) | |
| MSD | 2024 | ≥18 | NR | NHL | NR | NR | NR | |
| Dreger et al (2019)16,*,† | ||||||||
| Haplo | 132 | 58 (20-75) | 65 | DLBCL | 100/0 | 75/25 | PT-Cy ± CNI/MMF | 49 (12-73) |
| MSD | 525 | 55 (19-73) | 62 | DLBCL | 100/0 | 2/98 | CNI + (MMF 36%, MTX 45%, other 19%) | 48 (2-97) |
| Gauthier et al (2018)18,*,† | ||||||||
| Haplo | 61 | 29 (17-68) | 54 | HL | 100/0 | 51/49 | PT-Cy + CNI/MMF | 24 (3-58) |
| MSD | 90 | 32 (12-67) | 63 | HL | 100/0 | 14/86 | CNI + (none 39%, MMF 35%, MTX 25%) | 24 (3-70) |
| Ghosh et al (2016)14,*,† | ||||||||
| Haplo | 180 | 55 (18-75) | 64 | HL, NHL | 100/0 | 93/7 | PT-Cy ± CNI/MMF | 37 (6-73) |
| MSD | 807 | 54 (18-77) | 61 | HL, NHL | 100/0 | 2/98 | CNI + (MMF 31%, MTX 55%, other 14%) | 36 (3-76) |
| Martinez (2017)12 *,† | ||||||||
| Haplo | 98 | 31 (18-68) | 57 | HL | 90/10 | 61/39 | PT-Cy + CNI + (MMF 94%, other 6%) | 27 (1-64) |
| MSD | 338 | 32 (18-67) | 43 | HL | 69/31 | 10/89 | CNI + (MMF 27%, MTX 39%, other 34%) | 27 (1-76) |
| Robinson et al (2018)15,*,†,‡ | NR | |||||||
| Haplo | 218 | 41 (18-55) | 59 | AML, ALL | 40/60 | 57/43 | PT-Cy + CNI/MMF | |
| MSD | 843 | 42 (18-55) | 52 | AML, ALL | 16/84 | 12/88 | CNI + (MMF 20%, MTX 64%, none 16%) | |
| Robinson et al (2018)15,*,†,§ | NR | |||||||
| Haplo | 218 | 63 (55-76) | 59 | AML, ALL | 72/28 | 70/30 | PT-Cy + CNI/MMF | |
| MSD | 864 | 63 (55-76) | 58 | AML, ALL | 73/27 | 6/94 | CNI + (MMF 38%, MTX 45%, none 18%) | |
| Solh et al (2016)13 | 46 (12-123) | |||||||
| Haplo | 128 | 50 (19-74) | 54 | Any | 59/41 | 52/48 | PT-Cy + Tac/MMF | |
| MSD | 198 | 53 (18-77) | 59 | Any | 47/53 | 2/98 | Tac/MTX 66%, Tac/MMF 25%, other 9% |
The study by Bashey et al21 used the National Institutes of Health consensus criteria for cGVHD grading (mild vs moderate vs severe), whereas all other studies used the original classification system (limited vs extensive).
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; BM, bone marrow; CNI, calcineurin inhibitor; DLBCL, diffuse large B-cell lymphoma; F/U, follow-up; Haplo, haploidentical; HL, Hodgkin lymphoma; MA, myeloablative; MMF, mycophenolate mofetil; MTX, methotrexate; NHL, non-Hodgkin lymphoma; NR, not reported; PB, peripheral blood; PT-Cy, posttransplant cyclophosphamide; RI, reduced intensity; Tac, tacrolimus.
Multicenter study.
Registry study.
Recipient age 18 to 54 years.
Recipient age ≥55 years.
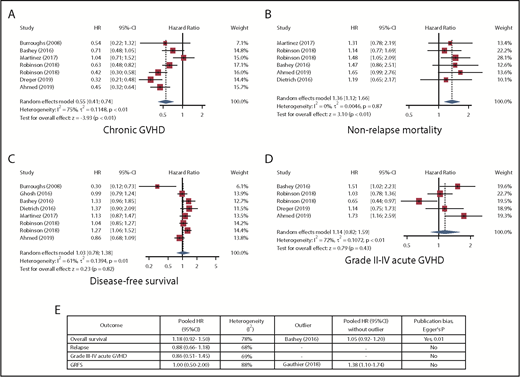
All studies were retrospective, 9 were multicenter, and all but 1 were published between 2016 and 2019. Meta-analysis results are shown in Figure 1. The most remarkable result was for cGVHD (7 studies; no outliers), with a pooled hazard ratio of 0.55 (95% confidence interval, 0.41-0.74) for haplo-HCT vs MSD, corresponding to ∼50% lower risk after haplo-HCT. In contrast, there was an increased risk for NRM (6 studies; no outliers) after haplo-HCT, with a pooled HR of 1.36 (95% confidence interval, 1.12-1.66). Results for OS (7 studies), DFS (8 studies), relapse (7 studies), GRFS (4 studies), and grade II-IV (5 studies) or III-IV (5 studies) acute GVHD were not significant. Egger’s regression suggested the presence of publication bias for OS (P = .01) and DFS (P = .07) but not for the other outcomes. All studies were classified as high quality (supplemental Table 1).
Random-effects meta-analysis. Detailed results for cGVHD (A), NRM (B), DFS (C), and grade II-IV acute GVHD (D). (E) A summary of other outcomes. The study by Robinson et al15 was considered separately for patient groups < 55 years of age (first occurrence in each plot) and ≥55 years of age (second occurrence in each plot) years. CI, confidence interval; HR, hazard ratio.
Random-effects meta-analysis. Detailed results for cGVHD (A), NRM (B), DFS (C), and grade II-IV acute GVHD (D). (E) A summary of other outcomes. The study by Robinson et al15 was considered separately for patient groups < 55 years of age (first occurrence in each plot) and ≥55 years of age (second occurrence in each plot) years. CI, confidence interval; HR, hazard ratio.
In the absence of a randomized controlled trial, our findings from a meta-analysis of several large studies indicate a consistent and substantially reduced risk for cGVHD after haplo-HCT compared with MSD. Grading of cGVHD and the chosen definitions were not consistent across studies. Of the 7 studies included for cGVHD analysis, 6 used the original classification (limited vs extensive),22 and 1 used the National Institutes of Health consensus criteria (mild vs moderate vs severe)23 for grading. However, the large magnitude of reduction in cGVHD after haplo-HCT suggests at least partial robustness to this inconsistency. Another limitation of our analysis is related to registry studies (n = 8). Although registry studies typically include large sample sizes, the quality of data depends on the reports from individual centers and may be variable. The reduction in cGVHD did not translate into more relapse; however, we found an increased risk for NRM after haplo-HCT. The reduction in cGVHD, the insignificant results for acute GVHD, and the separation of NRM curves early after HCT in most of the examined studies suggest infections and complications of delayed immune reconstitution as the main suspects for increased NRM after haplo-HCT, highlighting the value of infection-focused comparative studies and better supportive care.
Our analysis considered PT-Cy–based haplo-HCT and MSD without PT-Cy as “packages” and is not able to address the independent role of other relevant factors, such as conditioning intensity, graft source, and underlying disease. The preferred donor type may depend on these factors and their interactions, as suggested in recent works.15,24 Studies comparing haplo-HCT vs MSD, both using PT-Cy, as well as studies comparing MSD with vs without PT-Cy, could address the relative role of PT-Cy and HLA disparity in outcome differences observed in this meta-analysis. The results of 1 such study (BMT CTN 1203) were recently reported; a PT-Cy GVHD prophylaxis platform was the only 1 of the 3 randomized platforms that was superior to a nonrandomized contemporaneous tacrolimus plus methotrexate cohort for GRFS and cGVHD requiring immunosuppression in reduced-intensity matched-donor peripheral blood HCT.25 The BMT CTN 1703/1801 trial is comparing PT-Cy (plus tacrolimus and methotrexate) with tacrolimus plus methotrexate in reduced-intensity matched-donor peripheral blood HCT.
The full-text version of this article contains a data supplement.
Acknowledgment
The authors thank Todd E. DeFor and Xianghua Luo for statistical input.
Authorship
Contribution: A.R. designed the study and wrote the manuscript; M.A.M. conducted the literature search; M.A.M. and W.C. extracted the data; W.C. and H.C. performed the meta-analysis; L.L., A.B., X.Z., R.R., W.S., M.H., and D.J.W. critically evaluated the results and the manuscript; and A.B. and X.Z. provided additional data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Armin Rashidi, Division of Hematology, Oncology, and Transplantation, Department of Medicine, University of Minnesota, 14-132 Phillips Wangensteen Building, 516 Delaware St SE, Minneapolis, MN 55455; e-mail: arashidi@umn.edu.
References
Author notes
M.A.M. and W.C. contributed equally to this study.