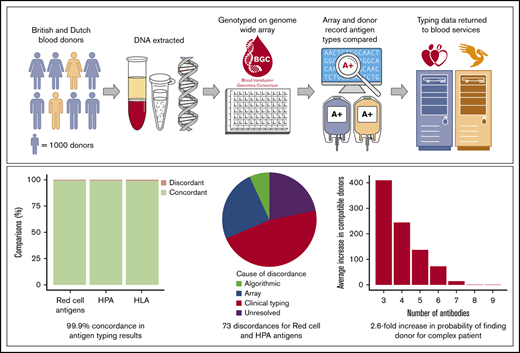
Key Points
Red blood cell, platelet, and white cell antigens can be typed accurately with a single, unified, DNA-based test.
As this test will be embedded in national genotyping studies, full blood cell typing data will be available for millions of individuals.
Abstract
Each year, blood transfusions save millions of lives. However, under current blood-matching practices, sensitization to non–self-antigens is an unavoidable adverse side effect of transfusion. We describe a universal donor typing platform that could be adopted by blood services worldwide to facilitate a universal extended blood-matching policy and reduce sensitization rates. This DNA-based test is capable of simultaneously typing most clinically relevant red blood cell (RBC), human platelet (HPA), and human leukocyte (HLA) antigens. Validation was performed, using samples from 7927 European, 27 South Asian, 21 East Asian, and 9 African blood donors enrolled in 2 national biobanks. We illustrated the usefulness of the platform by analyzing antibody data from patients sensitized with multiple RBC alloantibodies. Genotyping results demonstrated concordance of 99.91%, 99.97%, and 99.03% with RBC, HPA, and HLA clinically validated typing results in 89 371, 3016, and 9289 comparisons, respectively. Genotyping increased the total number of antigen typing results available from 110 980 to >1 200 000. Dense donor typing allowed identification of 2 to 6 times more compatible donors to serve 3146 patients with multiple RBC alloantibodies, providing at least 1 match for 176 individuals for whom previously no blood could be found among the same donors. This genotyping technology is already being used to type thousands of donors taking part in national genotyping studies. Extraction of dense antigen-typing data from these cohorts provides blood supply organizations with the opportunity to implement a policy of genomics-based precision matching of blood.
Introduction
The European Blood Alliance collects 31 million units of blood each year to provide life-saving support to an estimated 15 million individuals with a wide range of medical conditions.1 It is common practice to match red blood cells (RBCs) only for the ABO and RhD groups to ensure transfusion safety and prevent the majority of fatal hemolytic transfusion reactions (HTRs). However, sensitization to non–self RBC antigens remains an unavoidable consequence of this matching strategy.2-4
Annually, an estimated 3% (0.5 million) of patients become sensitized to RBC antigens after a single transfusion episode, with 60% of patients who receive regular transfusions becoming immunized.5-10 Sensitization confers a lifetime risk of HTRs, which from 2013 through 2017 were responsible for 17% (32 of 185) and 6% (7 of 110) of transfusion-related deaths reported to the US Food and Drug Administration and Serious Hazards of Transfusion UK, respectively.11,12 Sensitization can render transfusion-dependent patients nontransfusable and cause hemolytic disease in pregnancy, which is potentially life threatening to the fetus. Notwithstanding these serious side effects, the introduction of a more precise matching policy is resisted because of perceived logistical challenges and donor typing costs.13
Antibody-based tests are the current gold standard for RBC antigen typing; however, reliable reagents and high-throughput techniques are not available for all clinically relevant antigens. DNA-based tests have been used to overcome these limitations, and a range of in-house and commercial assays have been developed for donor genotyping.14-16 Studies have shown that antigen-negative blood can be supplied for 99.8% (5661/5672) of complex blood requests by using 43 066 donors genotyped for a limited number of RBC antigens.17 Despite this evidence most global blood supply organizations have not genotyped large numbers of their blood donors. The main reasons for this lack of uptake are the cost of current assays, the fact that no existing test can type all clinically relevant RBC antigens, and the lack of an algorithm for automated interpretation of results. Furthermore, existing tests do not include typing for other transfusion-relevant antigens, such as human leukocyte and platelet antigens (HLAs and HPAs), which are necessary for supporting cancer patients.18,19
A universal donor-typing platform must identify all clinically relevant RBC antigens for blood transfusions and HLAs and HPAs for platelet transfusions. The physical test must be combined with software for automated data interpretation and formatting so that it is immediately usable by blood supply organizations. Importantly, the platform must be cost effective and scalable to millions of donors and patients.
In earlier studies, we used whole-genome sequencing (WGS) and whole-exome sequencing for comprehensive RBC and HPA typing of patients, but sequencing has remained too costly to apply to vast numbers of blood donors.20,21 The Blood transfusion Genomics Consortium was established to capitalize on array technology recently applied in studies to genotype millions of individuals worldwide.22-25 In this report, we describe the validation of a high-throughput, genome-wide test repurposed for extensive blood donor antigen typing, which is available at a cost of ∼$40 per sample, inclusive of equipment, labor cost, and analysis.
Methods
Study design
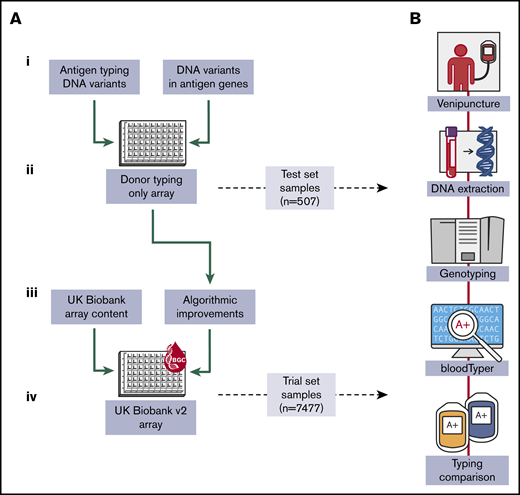
This study involved several interrelated components (Figure 1). First, we collated knowledge on 1602 known DNA sequence determinants of blood cell antigens.26-28 In an attempt to include genetic variants that may become important to antigen typing in the future, we also collated 9180 coding variants in 48 genes relevant to antigen expression (supplemental Table 1), all of which had minor allele frequencies >0.02% in large-scale sequencing data sets.29-32 Second, we fabricated an array containing this donor typing content and genotyped a small sample to detect any manufacturing or technical errors. Third, we integrated donor-typing content into the larger UK BioBank array design. The resulting Applied Biosystems UK Biobank version 2 Axiom Array (UKBBv2 array) includes content for both genome-wide typing and all currently known antigen-coding variants in RBC antigen- and HPA-encoding genes (Figure 2). Finally, the UKBBv2 array was tested on samples and data from thousands of blood donors by assessment of concordance between genetically and clinically determined antigen types. We also illustrated the advantages of donor genotyping by using referral data from difficult-to-match patients and a case report.
Overall project workflow. (A) Significant stages of the project. (i) Variants underpinning antigen expression and those in antigen-encoding genes in sequence variation databases were collected for incorporation into an array design. (ii) An array was fabricated containing only donor-typing content, and the small test set of donor samples was used to detect manufacturing or technical errors. (iii) Based on the results of the previous step, algorithmic changes were made to improve variant and antigen calling and donor-typing content was integrated into the UK Biobank Axiom array design. (iv) The resulting UK Biobank version 2 array was fabricated and the large trial set of donor samples was used to assess antigen typing performance. (B) Typical array testing cycle.
Overall project workflow. (A) Significant stages of the project. (i) Variants underpinning antigen expression and those in antigen-encoding genes in sequence variation databases were collected for incorporation into an array design. (ii) An array was fabricated containing only donor-typing content, and the small test set of donor samples was used to detect manufacturing or technical errors. (iii) Based on the results of the previous step, algorithmic changes were made to improve variant and antigen calling and donor-typing content was integrated into the UK Biobank Axiom array design. (iv) The resulting UK Biobank version 2 array was fabricated and the large trial set of donor samples was used to assess antigen typing performance. (B) Typical array testing cycle.
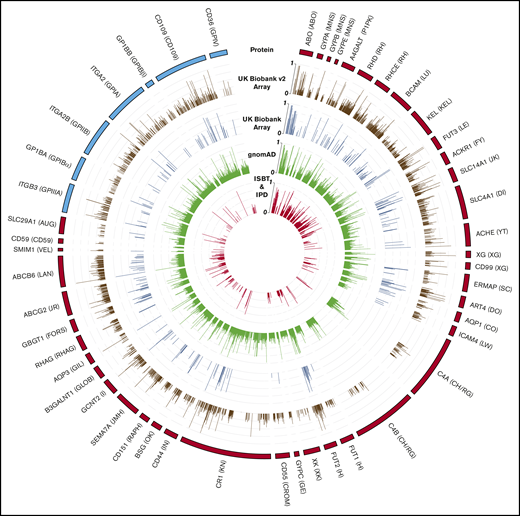
Genetic variation in antigen-controlling genes compared with array content. Tracks (1 to 5) from outside to inside: (1) representations of HPA (blue)- and RBC antigen (red)–encoding genes; box length equates to the number of amino acids in the protein. RBC and HPA system names are annotated next to gene labels; (2 and 3) coding variants included on the UK Biobank (UKBB) v2 (brown) and UKBB (dark blue) array, respectively; (4) all coding variants with consequences in the gnomAD database (green); (5) antigen-encoding variants in the ISBT and IPD-HPA databases (red). Bar length for tracks 2 to 5 represents gnomAD alternate allele frequency.
Genetic variation in antigen-controlling genes compared with array content. Tracks (1 to 5) from outside to inside: (1) representations of HPA (blue)- and RBC antigen (red)–encoding genes; box length equates to the number of amino acids in the protein. RBC and HPA system names are annotated next to gene labels; (2 and 3) coding variants included on the UK Biobank (UKBB) v2 (brown) and UKBB (dark blue) array, respectively; (4) all coding variants with consequences in the gnomAD database (green); (5) antigen-encoding variants in the ISBT and IPD-HPA databases (red). Bar length for tracks 2 to 5 represents gnomAD alternate allele frequency.
Blood donor samples
DNA samples were collected from 7984 blood donors in England and The Netherlands; 507 donors enrolled in the National Institute for Health Research (NIHR) BioResource were used as a “test set” to detect technical errors in the array fabrication process or data-processing software. The remaining 7477 samples, from blood donors in England (n = 4795) and The Netherlands (n = 2682) who had consented to participate in the COMPARE and DIS-III studies, were used as a “trial set” to validate the UKBBv2 donor genotyping array. Participant enrollment, ethics, sample processing, and DNA extraction methodologies used by the NIHR BioResource, COMPARE, and DIS-III studies have been reported.33,34
Genotyping and genotype QC
Each DNA sample was genotyped by using the Axiom genotyping platform at the Microarray Research Services Laboratory (Santa Clara, CA), in accordance with Axiom Best Practice Workflows.35 Total DNA of 750 ng at 30 ng/µL was used from each sample.
Genotypes were called using the AxiomGT1 algorithm included with Applied Biosystems Array Power Tools v2.10.2 software.36 Quality control (QC) metrics for each plate, sample and probe set were calculated automatically with the software. In brief, this involves analyzing the fraction of AT probe sets for which fluorescence is 2 standard deviations outside QC probe set fluorescence and assessing genotype call rates for each sample and variant. Probe sets failing the recommended thresholds were not used for analysis. For variants with more than 1 probe set remaining, a single best probe set was selected by using Fisher’s linear discriminant for cluster resolution.
To ensure that novel blood-typing content did not interfere with historical population-screening array content, we compared array-wide measured allele frequencies for 4795 COMPARE European-ancestry trial set donors to those measured in WGS data of 8511 European-ancestry participants in the NIHR BioResource pilot phase of the 100 000 Genomes Project (supplemental Figure 1).32 We also visually inspected genotype call plots of all newly added antigen-typing probe sets, per the original UKBB study methodology (supplemental Figure 2).22 Data generated by any poorly performing probe sets were not used for further analysis (supplemental Table 2).
DNA-based antigen typing
After QC was completed, genotype data were converted to Variant Call Format v4.2, using a combination of in-house software and BCFtools v1.3.1.37,38 Array-based RBC and HPA typing was then performed with the previously published bloodTyper algorithm modified for application to array data.39 HLA types were imputed from the array data with the freely available Applied Biosystems HLA Analysis v1.1 tool, by using the HLA*IMP:02 HLA type imputation model and multipopulation reference panel.40,41
Clinical antigen typing
Antigen typing data for the 7984 donors were generated by using clinically accredited tests as part of routine donor typing by National Health Service Blood and Transplant (NHSBT) and Sanquin. Only antigen types that had been verified by at least 2 independent measurements were used. We refer to these antigen types as “clinical types” as they are a combination of antibody- and DNA-determined types. Results were available for 48 RBC, 11 HPA, and 6 HLA antigens (supplemental Table 3).
Clinical typing results for ABO, D, C, c, E, e, K, k, Kpa, Jka, Jkb, Fya, Fyb, Lea, Leb, Lua, M, N, S, and s antigens were produced primarily using commercial high-throughput serological phenotyping systems (the Olympus PK7300 instrument, for example). Clinical types for the other RBC antigens were produced using in-house manual antibody-based typing tests. In-house polymerase chain reaction (PCR)–based techniques were used to type antigens such as Doa, Dob, and LAN, where no reliable antibody testing reagents are available. HLA typing results were produced with commercially available genotyping tests or in-house next-generation sequencing tests. HPA genotyping was performed with in-house–developed PCR-based tests.
RBC antigen and HPA typing concordance analysis
Antigens for which fewer than 10 comparisons between clinical and array antigen types were possible were excluded from the concordance analysis (supplemental Table 3). We also excluded the Lea and Leb antigens, because anti-Le antibodies are not usually clinically significant and the International Society of Blood Transfusion (ISBT) table required for variant interpretation is lacking.26,27 P1 antigen typing was disabled in bloodTyper, because the molecular basis of this antigen was not defined at the time of platform design.42
HLA typing concordance analysis
Four-digit HLA types were imputed from UKBBv2 array data by using the Applied Biosystems HLA Analysis v1.1 algorithm, which uses the HLA type-imputation model, HLA*IMP:02, and a multipopulation reference panel. Imputed HLA types were then compared with clinical HLA typing data extracted from NHSBT and Sanquin databases, with the following match algorithm:
Allele match: both results match for the first 2 fields (eg, HLA-A*32:01 and HLA-A*32:01:01).
String match: the clinical typing result is an ambiguous string of “potential” alleles that contains the genotype-inferred result (eg, HLA-A*02:01 is within HLA-A*02:01/HLA-A*02:04/HLA-A*02:07/HLA-A*02:09).
Group match: both results match in the first field, but because of the lack of clinical typing data, a second field comparison cannot be made (eg, HLA-A*24:02 and HLA-A*24).
Mismatch: both results are from different allele groups (eg, HLA-A*25:01 and HLA-A*26:01:01).
Targeted next-generation sequencing of discrepant samples
A targeted capture-sequencing assay was developed to resolve discordance between clinical and array antigen typing results. The assay design included all consensus coding sequences and 5′ and 3′ untranslated regions present in Ensembl and RefSeq for all relevant RBC antigen- and HPA-encoding genes. In addition, 1000 bp upstream of the transcript start sites were targeted. In total, 1 094 363 bp were targeted.
Aliquots of original DNA samples were retrieved from research biobanks, and 1 μg of each sample was fragmented with the E220 ultrasonicator (Covaris, Inc, Woburn, MA), to obtain average-sized fragments of 150-bp. Samples were processed using the TruSeq DNA LT Prep kit (Illumina Inc, San Diego, CA). Two DNA libraries were captured at the same time with a single reaction of the SeqCap BG capture array (Roche NimbleGen, Inc, Madison, WI). Enrichment was tested by quantitative PCR, measuring the abundance of 4 control target regions before and after DNA capture. The libraries were quantified by using the library quantification method (KAPA Biosystem, Ltd, Cape Town, South Africa) and sequenced by the Cancer Research UK Cambridge Institute Core Genomics facility in pools of 24 samples in 1 Illumina HiSeq 2000 lane, by 150-bp paired-end sequencing. QC and analysis of targeted sequencing data were performed with the previously published bloodTyper exome-sequencing workflow21 and visual review of sequencing alignments and variant calls by a panel of experts.
Results
Genotyping in this study was performed with 2 arrays of sequential design and 2 separate sets of donor samples. First, an array was fabricated that contained only donor-typing content and was evaluated by using 507 DNA samples from the test set for detection of manufacturing or technical errors. Second, donor-typing content was integrated into the UKBB array design, producing the UKBBv2 array, which was assessed in the 7477 DNA samples from the trial set.
Test array performance and Rh antigen typing algorithm
Concordance between clinical and array antigen-typing results for the test set was 87.17% in 7315 comparisons across 19 RBC antigens for which clinical typing was available (supplemental Figure 3). This analysis identified antigens, predominantly those of the Rh system, for which the quality of genotyping was unacceptably poor. The discordance was present for 2 reasons: first, the most common reason for the D-negative phenotype in Europeans is deletion of the entire RHD gene, and the initial version of the array did not detect the RHD copy number accurately. Second, genotyping of Rh antigens is known to be challenging because of high sequence homology between the RHD and RHCE genes.26 We made changes to array content and developed algorithms to overcome these difficulties (Figure 3). First, we tiled the RHD gene with 114 probe sets at loci with the lowest sequence homology to the RHCE gene. These probes allowed us to accurately identify the RHD copy number by using the standard CNVmix algorithm (Figure 3A-B).35 Second, we used the array-determined RHD copy number to segregate samples in 3 groups to overcome the effect of signal interference caused by cross-binding of RHCE probes to the RHD gene (Figure 3A,C). The application of this algorithm significantly increased concordance in the test set to 99.99%, 98.71%, and 99.61%, for the D, C/c, and E/e Rh group antigens, respectively (Figure 3D).
Novel Rh antigen-typing workflow. (A) Diagrammatic representations of the RHD and RHCE genes, the binding positions of array probes are indicated in red. The probes spanning RHD are used for RHD copy number assessment and for RHCE: for example, the probe for typing variant NM_020485.5:c.307C>T, which is used for C/c typing. (B) Results obtained for 2682 Dutch donor samples binned by RHD median copy number/probe set intensity ratio (log2), demonstrating that clear resolution of RHD copy number for each individual can be achieved by using the array. (C) Genotype call plot for the C/c antigen variant NM_020485.5:c.307C>T in the RHCE gene. Samples are colored by array-determined RHD copy number to allow visualization of the intensity shift caused by cross-binding of RHCE probes to RHD. Segregating samples in this way allows for accurate genotypes to be called. (D) Improvement in Rh(C/c) and Rh(E/e) antigen-typing concordance resulting from application of the novel genotyping algorithm.
Novel Rh antigen-typing workflow. (A) Diagrammatic representations of the RHD and RHCE genes, the binding positions of array probes are indicated in red. The probes spanning RHD are used for RHD copy number assessment and for RHCE: for example, the probe for typing variant NM_020485.5:c.307C>T, which is used for C/c typing. (B) Results obtained for 2682 Dutch donor samples binned by RHD median copy number/probe set intensity ratio (log2), demonstrating that clear resolution of RHD copy number for each individual can be achieved by using the array. (C) Genotype call plot for the C/c antigen variant NM_020485.5:c.307C>T in the RHCE gene. Samples are colored by array-determined RHD copy number to allow visualization of the intensity shift caused by cross-binding of RHCE probes to RHD. Segregating samples in this way allows for accurate genotypes to be called. (D) Improvement in Rh(C/c) and Rh(E/e) antigen-typing concordance resulting from application of the novel genotyping algorithm.
Large-scale, blinded trial of the UKBBv2 array
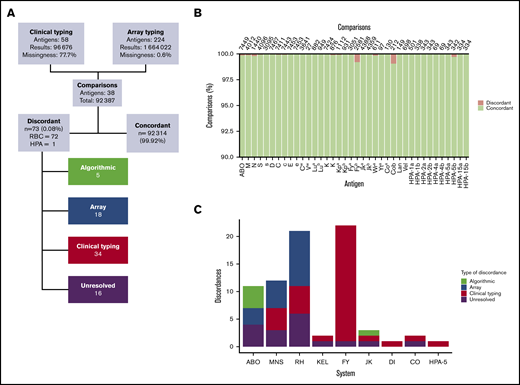
QC of 7477 trial set samples identified 15 samples with imputed vs declared sex mismatches and more than 5 antigen-typing discordances. Further investigation revealed that these discrepancies were caused by either erroneous handling of samples (n = 4) or incorrect data entry in the study database (n = 11). The data from the former category were excluded from further analysis, and the latter were retained in the study after error correction (supplemental Table 4), leaving 7473 samples in the final trial set. Overall concordance between clinical and UKBBv2 antigen typing was 99.82% in 103 326 comparisons across 28 RBC antigens, 10 HPAs, and 6 HLA loci for which clinical typing data were available (Figure 4; supplemental Table 3).
Concordance between clinical and UKBBv2 array antigen typing. (A) Overview of concordance analysis and high-level categorization of results. (B) Concordance per antigen is shown as a percentage of the total number of comparisons (given at the top of each bar) with concordant and discordant results in green and red, respectively. (C) Categorization by error type of discordances for the different RBC antigen systems and for the HPA-5 system.
Concordance between clinical and UKBBv2 array antigen typing. (A) Overview of concordance analysis and high-level categorization of results. (B) Concordance per antigen is shown as a percentage of the total number of comparisons (given at the top of each bar) with concordant and discordant results in green and red, respectively. (C) Categorization by error type of discordances for the different RBC antigen systems and for the HPA-5 system.
All but 57 of the 7473 trial set donors enrolled in this study were of European ancestry. Of the non-European individuals, 27, 21, and 9 were of South Asian, East Asian, and African ancestries, respectively. Antigen-typing concordance for these 57 individuals was 100% in 835 comparisons across the 28 RBCs and 10 HPAs compared.
RBC antigen-typing performance
Concordance between RBC antigen-typing results was 99.91% in 89 371 comparisons across 28 antigens (Figure 4A-B). We categorized the 72 (0.08%) remaining results according to the reason for discordance (Figure 4A,C; supplemental Table 5). Discordances (n = 33; 45.8%) were explained by erroneous clinical typing results. In 19 of these cases, variants encoding variant antigen expression were detected by the array, examples include Del (RHD*11, NM_016124:c.885T), Kmod (KEL*02M.01, NM_000420.2:c.1088G>A), and Fyx (FY*02W.01, NM_001122951.2:c.265C>T, and NM_001122951.2:c.298G>A). For carriers of these alleles, the chances of false-negative antibody-based typing results are greatly increased. A unit of blood that is erroneously typed as negative for a given antigen may boost antibody levels in a previously sensitized patient.43 The remaining 14 discordances in this category involved antigens for which current typing reagents have been known to give incorrect results (n = 4) or for which sequencing analysis confirmed array genotyping results (n = 10).
Sixteen of the discordances remain unresolved (supplemental Table 5). However, in 6 of these cases, previously unobserved DNA variants, which were likely to underpin an antigen-negative phenotype were discovered by using the targeted sequencing platform designed for discordance resolution. The absence of these newly identified variants from the ISBT reference tables prevented their use in antigen phenotype inference. To determine the effect of these 6 unique variants on antigen expression, functional studies are required; therefore, we regard these cases as unresolved. For the remaining 10, we were unable to resolve the cause of the discordance because of the lack of samples for further analysis.
HLA typing
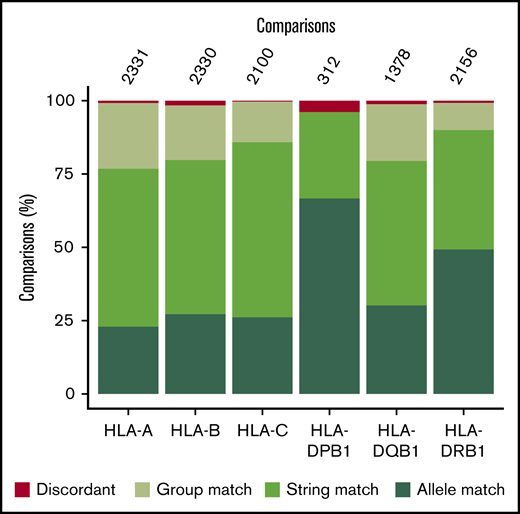
Most blood supply organizations maintain panels of HLA class I–typed apheresis platelet donors to support patients who are refractory to ABO-compatible platelet concentrates related to HLA class I antibodies. Clinical HLA typing data were available for 1221 of the 7473 trial set donors. Concordance between typing results across 6 HLA class I and II loci, at 2-field resolution, was 99.03% in 9289 comparisons (Figure 5; supplemental Table 6).
Concordance between clinical and UKBBv2 array HLA-typing results. Concordance per antigen is shown as a percentage of the total number of comparisons (given at the top of each bar) with concordant and discordant results in green and red, respectively. The concordance was assessed at group, string, and allele levels.
Concordance between clinical and UKBBv2 array HLA-typing results. Concordance per antigen is shown as a percentage of the total number of comparisons (given at the top of each bar) with concordant and discordant results in green and red, respectively. The concordance was assessed at group, string, and allele levels.
HPA typing
A portion of patients with HLA class I antibodies also form HPA antibodies. For these patients, platelet concentrates lacking the relevant class I HLAs and HPAs are required. We report 99.96% concordance in 3017 comparisons across 10 antigens, with only a single discordant result observed, for HPA-5a (Figure 4; supplemental Tables 3 and 5).
Impact of an extensively genotyped donor panel
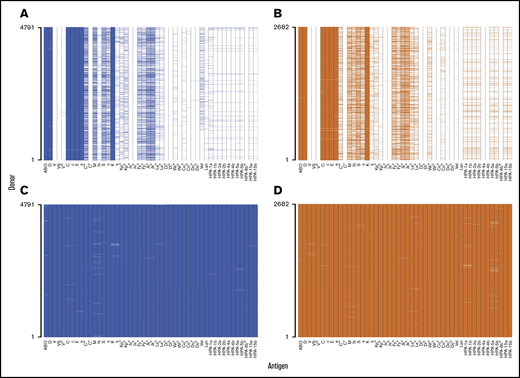
For 7473 trial-set donors 37 RBC antigens and 11 HPAs had at least 1 clinical typing result available. These 48 antigens formed a data set that was representative of extended donor typing programs conducted on British and Dutch donors (Figure 6A-B). In contrast to clinical typing data, genotype-inferred antigen typing was almost complete, with only 458 (0.13%) of 354 624 possible results missing (Figure 6C-D). Genotyping yielded 3.8 times more types (47.9 vs 12.6 results per donor) compared with current practice. Genotyping also allowed us to type the donors for 185 additional antigens for which no clinical typing results were available. In total, 1.2 million antigen types, on average 214.6 per donor, were produced by genotyping.
Comparison of clinical and UKBBv2 array antigen–typing availability in the trial set of samples. The presence of color represents a typing result (positive or negative), and the absence of color indicates lack of a typing result. Clinical and UKBBv2 array antigen typing data are shown for British (blue; A,C) and Dutch donors (orange; B,D).
Comparison of clinical and UKBBv2 array antigen–typing availability in the trial set of samples. The presence of color represents a typing result (positive or negative), and the absence of color indicates lack of a typing result. Clinical and UKBBv2 array antigen typing data are shown for British (blue; A,C) and Dutch donors (orange; B,D).
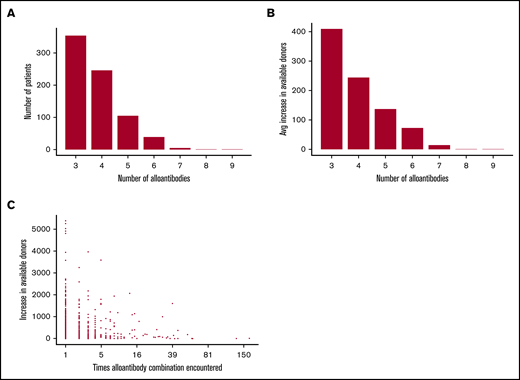
To investigate the potential benefits of densely genotyping donors we used data from patient referrals to NHSBT over a 5-year period. Filtering this data set for “complex cases,” defined as patients with at least 3 RBC alloantibodies, produced a set of 3146 referrals with 1253 unique alloantibody combinations (Figure 7A; supplemental Table 7). We observed an inverse relationship between the probability of finding a compatible donor and the number of alloantibodies identified in a patient (Figure 7B). Using the trial set of 7473 donors as a “virtual stock,” we report a 2.6-fold greater probability of identifying ABO- and RhD-compatible donors who are negative for the relevant combination of antigens needed to support patients with 1 of the 1253 alloantibody combinations, when using the genotype data. This translated to an additional 176 patients for whom a matched donor could be identified. When finding donors for the most common alloantibody profiles, genotyping data were equivalent to clinical typing data, but for the rare combinations, genotyping increased the average number of identified donors by 72 (Figure 7C). We were even able to identify a compatible donor for a patient with alloantibodies against 9 different RBC antigens.
UKBBv2 array genotyping significantly increases the number of available units for patients with multiple RBC antibodies. (A) The number of alloantibodies identified in patient samples, with the number of patients and antibodies on the vertical and horizontal axis, respectively. (B) Average increase in the number of compatible donors when using UKBBv2 array instead of clinical typing data to identify compatible donors, cataloged by the number of RBC alloantibodies identified in patient samples. (C) The increase in the number of compatible donors when using the UKBBv2 array instead of clinical typing data for each complex alloantibody profile identified in the data from 3146 patients with RBC alloantibodies.
UKBBv2 array genotyping significantly increases the number of available units for patients with multiple RBC antibodies. (A) The number of alloantibodies identified in patient samples, with the number of patients and antibodies on the vertical and horizontal axis, respectively. (B) Average increase in the number of compatible donors when using UKBBv2 array instead of clinical typing data to identify compatible donors, cataloged by the number of RBC alloantibodies identified in patient samples. (C) The increase in the number of compatible donors when using the UKBBv2 array instead of clinical typing data for each complex alloantibody profile identified in the data from 3146 patients with RBC alloantibodies.
Immediate clinical benefit of extended typing data
Illustrating the clinical need for denser typing of donors, we present a female patient with myelodysplastic syndrome with anti-E, -Coa, and -Wra antibodies. Because of her bone marrow failure, she required regular transfusions; however, no suitable donors were identified in the 341 509 Dutch donors registered with Sanquin. Querying genotyping results from the 2682 Dutch donors in this study identified 5 compatible active donors; their donations have now been used to provide life-saving transfusion support for this patient.
Donor and patient health information
Because of the large number of genetic variants typed by the UKBBv2 array, the utility of this platform extends far beyond antigen typing for extended matching. For example, homozygosity for the variant NM_000410.3:c.845G>A in the HFE gene is the most common cause of hereditary hemochromatosis. A recent study showed that 21.7% of male and 9.9% of female UKBB participants with this genotype presented with iron-overload pathologies.44 Using the UKBBv2 array data, we identified 78 (1%) donors in the trial set testing as homozygous for this variant. This information provides blood supply organizations with an opportunity to reduce the risk of such pathologies developing in these individuals by recommending frequent donations.
Discussion
To ensure transfusion safety, blood is routinely matched for ABO and RhD compatibility. However, this policy is not generally applied to other RBC antigens, and each year ∼0.5 million patients become sensitized as a consequence. These patients subsequently require extended matching of blood to prevent HTRs, which are occasionally lethal. This situation can become particularly precarious in transfusion-dependent patients, and it has been shown that extended matching from the beginning of transfusion support brings immediate benefits by reducing the incidence of alloantibody formation.45,46 To reduce the frequency of this serious hazard of transfusion, some blood supply organizations have introduced an extended matching policy for these patient groups. However, the cost of implementing generalized extended-matching policies will not be justified without conclusive evidence from randomized trials of consequent reductions in morbidity or mortality. For such trials to be possible, a comprehensive and affordable extended antigen-typing assay is needed, to test large numbers of donors and patients.
Herein, we report a technology that is already used to genotype millions of individuals and is now optimized for donor typing. We showed by genotyping 7473 donors, 99.92% concordance between clinical and array antigen typing results in 103 326 comparisons across 44 clinically relevant RBC antigens and HLAs and HPAs. We observed 1 HPA and 72 RBC discordances between clinical and genetically inferred antigen type (0.08% of comparisons).
Thirty-four discordances were caused by incorrect clinical typing results. In 22 of these cases, antigens were typed negative by antibody-based clinical typing, but were correctly inferred to be present by genotyping. Transfusion of such blood into patients who have been presensitized with the corresponding antibody is clinically unacceptable, as it may cause an HTR. For these cases, our data showed that genotyping was superior to clinical typing. Importantly, the array identified 2 individuals positive for the D antigen who were erroneously typed serologically as D-negative. Further analysis of genotyping and sequencing results revealed that these individuals possessed the Del phenotype (RHD*11 and RHD*01EL.09). Detection of variant Rh antigens such as Del is of particular importance when providing blood for transfusion-dependent patients. Although the array contains content to enable typing of all ISBT known Rh variant antigens, further validation using a larger number of donors typed for these antigens is needed.
Twenty-three discordant results were caused by an array genotype error (n = 18) or incorrect bloodTyper interpretation (n = 5). Although only 3 of these cases resulted in false-negative typing results, this error type may result in an incompatible transfusion if blood units are issued according to electronic cross-matching, using genotyping data alone. After these observations, we made modifications to the bloodTyper algorithm to reduce the chance that such errors would occur. However, 7 discordances involved ABO antigens. Because of the clinical severity of ABO- and RhD-mismatched blood, it remains international best practice, to type every donation for these antigens by using antibody-based methods.
The question of whether extended typing of blood donors improves the efficiency of matching blood units to patients with multiple RBC alloantibodies has been much debated. When extended genotyping data were made available, we observed a 2.6-fold increase in the number of donors identified to support 3146 complex patients with multiple alloantibodies. The immediate clinical benefit of having densely typed donors was demonstrated by a female patient with severe anemia related to bone marrow failure who required blood with an antigen-typing profile possessed by 1 in 200 donors. Notwithstanding the relatively high frequency, no suitable donors were present in The Netherlands, because donors are not routinely typed for Coa and Wra, because reliable or affordable typing reagents are lacking. Five compatible Dutch donors were identified in the trial set of this study.
We did not alter the UKBB array content at the HLA locus, because findings in previous studies suggest that class I and II HLAs can be imputed with good accuracy from the original UKBB array data.40 However the level of accuracy has never been empirically verified by direct comparison of array vs clinical grade HLA-typing data. We provide this comparison, which showed excellent concordance at a 99.03% accuracy at 2-field resolution. Typing at this resolution has immediate value for the provision of HLA class I–matched platelet concentrates for refractory patients and shows that the platform, if widely applied, could be used to improve the composition of platelet apheresis panels via identification of donors homozygous for rare HLA class I haplotypes.
We used the HFE locus to illustrate that the array data can also be used to inform policies about donor health and confirm that ∼1% of donors are homozygous for the hemochromatosis-causing HFE variant. Based on this observation, an estimated 12 000 individual donors in England and The Netherlands could donate more frequently, thereby reducing their risk of developing ill health while adding to blood stocks.
Reflecting the donor registries of The Netherlands and Britain, only 0.7% of donors available for this study were of non-European ancestry. Because of the low frequency of Rh variant antigens in Europeans and their high frequency in patients of African ancestry with hemoglobinopathy, further validation must be performed before this test can be used to clinically type these patients and the required donors for variant antigens. However, the complete concordance between 835 antigen-typing results for the 57 non-European individuals and the identification of 2 erroneous serological typing results caused by variant RhD expression reported herein provides support for further studies using samples from donors and patients recruited by ancestry and specific blood antigen types.
We present, for the first time, an affordable genotyping assay capable of simultaneously typing all clinically relevant human RBC group antigens, which has been trialed in thousands of blood donors. The test is available in 2 formats: a genome-wide–typing array that can be used in population-scale genotyping studies and a donor-typing array containing only content relevant to antigen typing that can be used where genome-wide typing is not an option. As the study was embedded in the national blood services of England and The Netherlands, we were able to take a first look at how high-volume comprehensive donor typing can be used to simplify provision of compatible blood for sensitized patients with alloantibodies against multiple RBC antigen groups. Uniquely, the data generated in this study are being used to provide life-saving transfusion support for a young female patient for whom no compatible blood was available. The success in this case highlights the immediate improvements in health care that this technology has to offer.
We also see opportunities for improvements in the medium term. With many health care systems making preparations for the era of precision medicine, an increasing number of countries are genotyping a proportion of their population. The assay developed in this study is nested within the platform already used for population-genotyping studies, such as the FinnGen Biobank, Million Veteran Program, Taiwan Precision Medicine Initiative, UK Biobank, and the impending UK 5 Million study. The genotyping in these studies will provide full blood cell antigen types on millions of individuals, giving blood supply organizations the opportunity to implement a policy of genomics-based transfusion medicine.
Genotype data of study participants from all sources can be retrieved from the European Genome-phenome archive (https://www.ebi.ac.uk/ega/home) at the European Molecular Biology Laboratory European Bioinformatics Institute (Hinxton, Cambridge, United Kingdom; EMBL-EBI) using the following accession numbers: NIHR BioResource participants (EGAD00001005024); COMPARE study participants (EGAD00001005023); DIS-III study participants (EGAD00001005026).
Acknowledgments
The Academic Coordinating Centre thanks the Blood Donor Centre staff and blood donors for participating in this study. This study made use of data generated by the COMPARE and DIS-III studies. Genotyping results for COMPARE participants were retrieved from the NIHR BioResource. The authors thank NIHR BioResource volunteers for their participation; NIHR BioResource Centres, NHS Trusts, and NHS Blood and Transplant (NHSBT) and staff for their contribution; the Locus Reference Genomic team at the EBI for their assistance in curating gene transcripts; and all colleagues, in particular, Alan Gray and Candice Davidson (NHSBT Tooting Centre, London, United Kingdom) for scientific and clinical advice.
Funding for the project was provided by NHSBT, NIHR (grant RG65966) and Sanquin (grant PPOC 14-028). The Academic Coordinating Centre for the COMPARE and INTERVAL studies was supported by core funding from the NIHR Blood and Transplant Research Unit (BTRU) in Donor Health and Genomics (grant NIHR BTRU-2014-10024), the UK Medical Research Council (MRC; grant MR/L003120/1), the British Heart Foundation (grants SP/09/002; RG/13/13/30194; RG/18/13/33946), and the NIHR (Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust). This work also made use of the UKBB resource under application 13745 and received support from Health Data Research UK, which is funded by the MRC, the Engineering and Physical Sciences Research Council, the Economic and Social Research Council, the Department of Health and Social Care (England), the Chief Scientist Office of the Scottish Government Health and Social Care Directorates, the Health and Social Care Research and Development Division (Welsh Government), the Public Health Agency (Northern Ireland), the British Heart Foundation, and the Wellcome Trust. Participants in the COMPARE and INTERVAL trials were recruited with the active collaboration of NHSBT, which supported field work and other elements of the trial. DNA extraction and genotyping was cofunded by the NIHR BioResource and the NIHR, via the Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust.
Complete lists of the investigators and contributors to the INTERVAL and COMPARE studies have been published (Lancet. 2017;390(10110):2360-2371 and ISRCTN Registry 2017;ISRCTN90871183). The views expressed are those of the authors and are not necessarily those of the NHS, NIHR, or the Department of Health and Social Care in England.
Authorship
Contributions: N.S.G., B.V., A.S.B., W.H.O., C.E.v.d.S., N.A.W., W.J.A., and W.J.L. designed the study and wrote the manuscript; N.S.G., B.V., J.G., J.O., C.J.P., C.M.S., N.v.d.B., K.M., C.S.N., S. Tuna, R.V., and W.J.L. analyzed data; E.D.A., T.C.T., K.v.d.H., D.D., J.L., I.S., J.D., D.J.R., and K.E.S. provided project support and organized participant recruitment; and C.B., K.B., A.T.D., S.G., J.M.J., G.M., F.J.P., J.S., M.S., N.T., S. Trompeter, M.R.W., A.R., and C.M.W. provided expert knowledge.
Conflict-of-interest disclosure: J.G., R.V., and C.S.N. are employees of Thermo Fisher Scientific. A.T.D. is a cofounder of Peptide Groove, LLP, a company that commercializes statistical HLA-typing approaches. The remaining authors declare no competing financial interests.
A complete list of the members of the Blood Transfusion Genomics Consortium appears in the supplemental appendix.
Correspondence: Nicholas S. Gleadall, Department of Haematology, Cambridge Biomedical Campus, University of Cambridge, Cambridge CB2 0XY, United Kingdom; e-mail: ng384@cam.ac.uk.
References
Author notes
N.S.G., B.V., and J.G. contributed equally to this study.
The full-text version of this article contains a data supplement.