TO THE EDITOR:
Chronic lymphocytic leukemia (CLL) is the most prevalent type of leukemia in adults in western countries. Family studies demonstrated that first-degree relatives of patients with CLL face a 6- to 12-fold increased risk of developing the disease and a 2-fold increased risk of developing another non-Hodgkin lymphoma.1 Familial CLL has generally been defined as a family with at least 2 first-degree relatives who are both affected with CLL.2 Genome-wide association studies identified several susceptibility loci, but despite these advances, molecular analysis for predisposition to CLL is still an ongoing subject of research because a vast majority of familial cases remain unexplained.3-5
Many efforts have been dedicated to the analysis of the heavy chain of the surface immunoglobulin and revealed that the mutational load within the immunoglobulin heavy variable (IGHV) genes distinguishes mutated CLL with a markedly superior prognosis from unmutated CLL cases.6 A recent study unveiled that the light chain can also affect CLL prognosis and that IGLV3-21 light chain usage in itself defines a new subgroup of patients with CLL with poor prognosis.7
In 2020, 2 studies provided the rationale for this observation, reporting the functional and clinical consequences of a somatic, single-point mutation in the IGLV3-21 gene, changing the glycine (G) at position 110 to an arginine (R), conferring autonomous B-cell receptor (BCR) signaling.8,9 This acquired mutation was associated with an adverse clinical outcome, independent of the IGHV status. Interestingly, it seems that only the IGLV3-21*04 allele (recently described allele, differing from the initially reported *01 allele by 1 nucleotide) holds the prerequisite BCR amino acid motifs required for the homotypic BCR–BCR interaction, triggering autonomous signaling and thus representing an inherited risk factor for CLL.10 Given the assumption that this allele constitutes an inherited risk factor for CLL, we assessed the prevalence of both the IGLV3-21*04 allele and the R110 mutation in a cohort of patients with familial CLL.
Forty-five patients with CLL from the Registre National des Hémopathies Lymphoïdes Familiales, with a familial history of CLL (n = 41) or other mature B-cell neoplasm (n = 4), were included in the study. They were selected on expression of the lambda light chain by flow cytometry from a total cohort of 130 patients. The study was approved by the Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale (CCPPRB) of Versailles. All samples were obtained with informed consent and used in full compliance with institutional regulations. Genomic DNA was extracted from peripheral blood (PB) samples using the QiAmp DNA mini kit (Qiagen, Hilden, Germany).
Detection of the IGLV3-21R110 mutation was performed using an in-house, cost-effective restriction fragment length polymorphism (RFLP) technique, enabling faster screening of large samples series compared with traditional sequencing. First, we designed polymerase chain reaction (PCR) primers allowing specific amplification of IGLV3-21–IGLJ1/2/3 introns rearrangements (other IGLJ genes were not targeted as very rarely used in CLL11,12 ). The forward primer is labeled with FAM fluorochrome and is specific for the IGLV3-21 gene. The 2 reverse primers are positioned in IGLJ introns to amplify the entire IGLJ genes, including the last nucleotide where the mutation occurs (forward primer: 5′-CATCAGCAGGGTCGAAGCC-3′; reverse primers: 5′-TACCCGGAGACTAATGCACC-3′ [J1 intron] and 5′CAAGCAAGGGTCTGAACAGG-3′ [J2-J3 intron]).
As the R110 mutation creates a de novo restriction site (A/CGT) for the HpyCH4IV enzyme (New England Biolabs), PCR products were digested 1 hour at 37°C with this restriction enzyme. After digestion, PCR products were analyzed by capillary electrophoresis (ABI 3730; Applied Biosystems). The presence of the R110 mutation results in a shorter fluorescent DNA fragment, identifiable on the profiles generated by GenemapperTM Software 5.
The performance of our experimental design was verified by several means. We first selected known sequences containing or not containing the restriction site. As expected, the HpyCH4IV enzyme only digested PCR products harboring the restriction site (data not shown). Because previous publications reported that 100% of subset 2 CLL harbor the R110 mutation,8,9 we collected 25 subset #2 CLL cases from our department and confirmed that all were digested after PB DNA amplification with our set of primers. Finally, we collected PB DNA from 33 non-subset #2 lambda CLL, from which 10 showed a clonal IGLV3-21 rearrangement. Those 10 cases underwent digestion, and presence (n = 6) or absence (n = 4) of the mutation was doubled-checked using Sanger Sequencing (supplemental Figure 1). The lack of PCR amplification of the remaining 23 cases was explained by the usage of IGLV1/IGLV2 subgroup genes (n = 14), IGLV3 other than IGLV3-21 (n = 2; confirmed by Sanger sequencing), or involvement of another IGLV/IGLJ gene (n = 7) in the rearrangement of the tumor cells (data not shown).
The study population included 45 patients, all from different families, selected on monotypic lambda light chain expression by reanalysis of immunophenotyping data. Families had 2, 3, 4, or 5 kindreds affected in 30 of 45 (66.7%) 9 of 45 (20%), 3 of 45 (6.7%), and 3 of 45 (6.7) of cases, respectively. Median age at diagnosis was 59 years old (range, 36-83 years), and the male/female ratio was 1.4. The screening of the lambda cases among the whole cohort (n = 130) indicated that, as reported in sporadic CLL, approximately one third of familial CLL express this light chain isotype.11,12
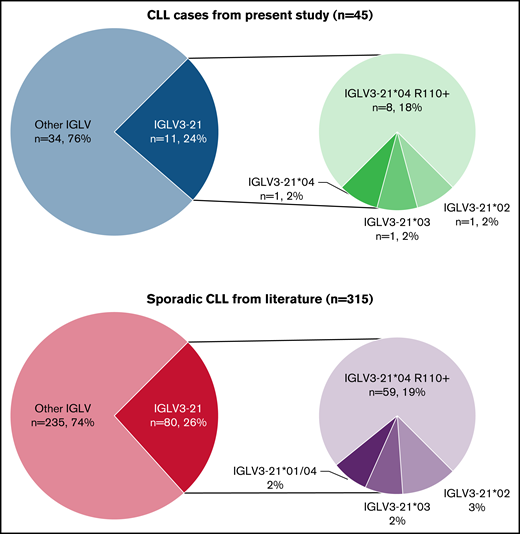
A clonal rearrangement using the IGLV3-21 gene was detected with our specific IGLV3-21 PCR assay for 11 of 45 cases (24%). Those 11 cases were further evaluated for the presence of the mutation by digestion with the HpyCH4IV restriction enzyme. Eight of 11 cases (73%) were positive for digestion (ie, harbored the mutation R110). We assessed that the lack of amplification of the 34 other cases was not due to a technical issue by performing the BIOMED-2 control PCR for assessment of amplifiability and integrity of DNA samples, which ruled out this hypothesis (data not shown).13
Sanger sequencing of the 11 IGLV3-21 cases indicated that all 8 R110-positive cases had the allele IGLV3-21*04, whereas the 3 R110-negative cases had IGLV3-21*02, IGLV3-21*03, and IGLV3-21*04, respectively.
Finally, we examined whether the CLL-affected relatives of the 8 R110-positive cases had the same light chain features. Flow cytometry data were available for 6 of them, revealing that only kappa-expressing CLL was found in the relatives (n = 7; Table 1; supplemental Figure 2).
Characteristics of the 11 familial lymphoproliferative disorder cases with IGLV3-21 gene usage
| Case . | Age at diagnosis, y . | Sex . | IGHV . | Subset 2 . | IGLV3-21 . | R110 . | Relatives with CLL (light chain isotype) . |
|---|---|---|---|---|---|---|---|
| 1 | 83 | Female | IGHV4-4 | No | Yes | Yes | Brother (kappa) |
| 2 | 65 | Female | IGHV3-21 | Yes | Yes | Yes | Mother (unknown) |
| 3 | 66 | Female | IGHV1-18 | No | Yes | Yes | Brother (kappa) |
| 4 | 48 | Male | IGHV3-48 | No | Yes | No | NA* |
| 5 | 49 | Male | IGHV3-23 | No | Yes | Yes | Brother (kappa); father (unknown) |
| 6 | 64 | Male | IGHV1-69 | No | Yes | No | NA* |
| 7 | 73 | Male | IGHV3-21 | Yes | Yes | Yes | Brother (kappa); daughter (kappa) |
| 8 | 48 | Male | Unknown | No | Yes | Yes | NA† |
| 9 | 55 | Male | IGHV3-15 | No | Yes | No | NA* |
| 10 | 65 | Male | IGHV4-59 | No | Yes | Yes | Cousin (kappa) |
| 11 | 40 | Female | IGHV3-66 | No | Yes | Yes | Mother (kappa) |
| Case . | Age at diagnosis, y . | Sex . | IGHV . | Subset 2 . | IGLV3-21 . | R110 . | Relatives with CLL (light chain isotype) . |
|---|---|---|---|---|---|---|---|
| 1 | 83 | Female | IGHV4-4 | No | Yes | Yes | Brother (kappa) |
| 2 | 65 | Female | IGHV3-21 | Yes | Yes | Yes | Mother (unknown) |
| 3 | 66 | Female | IGHV1-18 | No | Yes | Yes | Brother (kappa) |
| 4 | 48 | Male | IGHV3-48 | No | Yes | No | NA* |
| 5 | 49 | Male | IGHV3-23 | No | Yes | Yes | Brother (kappa); father (unknown) |
| 6 | 64 | Male | IGHV1-69 | No | Yes | No | NA* |
| 7 | 73 | Male | IGHV3-21 | Yes | Yes | Yes | Brother (kappa); daughter (kappa) |
| 8 | 48 | Male | Unknown | No | Yes | Yes | NA† |
| 9 | 55 | Male | IGHV3-15 | No | Yes | No | NA* |
| 10 | 65 | Male | IGHV4-59 | No | Yes | Yes | Cousin (kappa) |
| 11 | 40 | Female | IGHV3-66 | No | Yes | Yes | Mother (kappa) |
NA, not applicable.
Cases 4, 6, and 9, absence of R110 mutation.
Case 8, relative with follicular lymphoma.
Compared with sporadic CLL, our results are highly similar to those reported in different cohorts from the literature, with 21% to 26% of sporadic lambda chain CLL using the IGLV3-21 gene (24% in our cohort) and 71% to 89% of IGLV3-21 CLL cases harboring the R110 mutation (73% in our cohort).7-9
In addition, although the number of cases in our cohort is limited, we report a comparable prevalence of the IGLV3-21*04 allele in R110-positive cases (100% in both sporadic CLL and our cohort) and in R110-negative cases (25%-27% in sporadic CLL, 33% in our cohort; Figure 1).
Proportion of IGLV3-21 gene and allele usage, and R110 mutation in lambda-expressing CLL. (A) Results of the present cohort of 45 familial lymphoproliferative disorder cases (41 familial CLL, 4 families with CLL and another B-cell malignancy including 2 Hodgkin lymphoma, 1 follicular lymphoma, 1 nonspecified non-Hodgkin lymphoma). (B) Results from public datasets (pooled data from supplemental material published elsewhere8,9 ).
Proportion of IGLV3-21 gene and allele usage, and R110 mutation in lambda-expressing CLL. (A) Results of the present cohort of 45 familial lymphoproliferative disorder cases (41 familial CLL, 4 families with CLL and another B-cell malignancy including 2 Hodgkin lymphoma, 1 follicular lymphoma, 1 nonspecified non-Hodgkin lymphoma). (B) Results from public datasets (pooled data from supplemental material published elsewhere8,9 ).
We conclude that, in our cohort, the prevalence of IGLV3-21*04 and R110 mutation among individual familial cases of CLL does not differ from sporadic CLL. Furthermore, we did not observed clustering of cases within the same family. In this regard, a recent study reported a family where all 4 siblings had the IGLV3-21*04, R110-positive feature.14 However, the 7 family members we were able to further investigate in our cohort revealed a kappa light chain usage in the CLL population, indicating that the emergence of CLL in those cases was not attributable to the presence of the IGLV3-21*04 allele. However, further studies on large series are warranted to establish the definitive hereditary risk factor for CLL held by the IGLV3-21*04 allele.
As the R110 mutation has prognosis implications, the rapid and affordable technique we described in this work allows for systematic evaluation at CLL diagnosis, which can be easily implemented in parallel to IGHV mutation status assessment.
Acknowledgments: M.A. is supported by an ERA-NET TRANSCAN-2/French National Cancer Institute grant (NOVEL consortium). This work was supported in part by Cancer United Research Associating Medicine, University and Society (CURAMUS), INCA-DGOS-Inserm_12560.
Contribution: M.A. and F.D. designed the study; F.T. and S.C. collected and interpreted the clinical data; M.A. and P.V. performed the experiments; M.A., F.D., C.B., and M.L.G.-T. contributed to the data interpretation; M.A. and F.D. wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: S.C. served on the scientific advisory board for Gilead, Novartis, Roche France, Abbvie, Sandoz, Sanofi, Janssen, Celgene-BMS, Takeda, Atara. The remaining authors declare no competing financial interests.
Correspondence: Frédéric Davi, Laboratoire d'Hématologie Biologique, Hôpital Pitié-Salpêtrière, 47-83 Bd de l'Hôpital, 75013 Paris, France; e-mail: fred.davi@aphp.fr.
References
Author notes
Requests for data sharing may be submitted to Frédéric Davi (fred.davi@aphp.fr).
The full-text version of this article contains a data supplement.