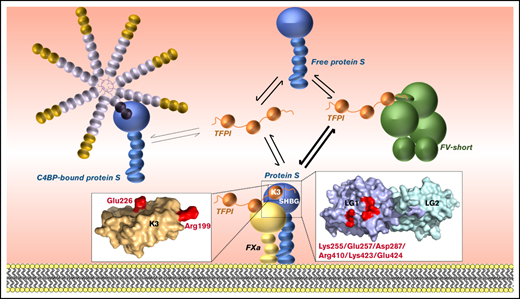
Key Points
Protein S LG1 residues Lys255/Glu257/Asp287/Arg410/Lys423/Glu424 are critical for its TFPI cofactor function.
Binding of the C4BP β-chain to protein S blocks the function of this region.
Abstract
Protein S is a cofactor in the tissue factor pathway inhibitor (TFPI) anticoagulant pathway. It enhances TFPIα-mediated inhibition of factor (F)Xa activity and generation. The enhancement is dependent on a TFPIα-protein S interaction involving TFPIα Kunitz 3 and protein S laminin G-type (LG)-1. C4b binding protein (C4BP), which binds to protein S LG1, almost completely abolishes its TFPI cofactor function. However, neither the amino acids involved in TFPIα enhancement nor the mechanisms underlying the reduced TFPI cofactor function of C4BP-bound protein S are known. To screen for functionally important regions within protein S LG1, we generated 7 variants with inserted N-linked glycosylation attachment sites. Protein S D253T and Q427N/K429T displayed severely reduced TFPI cofactor function while showing normal activated protein C (APC) cofactor function and C4BP binding. Based on these results, we designed 4 protein S variants in which 4 to 6 surface-exposed charged residues were substituted for alanine. One variant, protein S K255A/E257A/D287A/R410A/K423A/E424A, exhibited either abolished or severely reduced TFPI cofactor function in plasma and FXa inhibition assays, both in the presence or absence of FV-short, but retained normal APC cofactor function and high-affinity C4BP binding. The C4BP β-chain was expressed to determine the mechanisms behind the reduced TFPI cofactor function of C4BP-bound protein S. Like C4BP-bound protein S, C4BP β-chain-bound protein S had severely reduced TFPI cofactor function. These results show that protein S Lys255, Glu257, Asp287, Arg410, Lys423, and Glu424 are critical for protein S-mediated enhancement of TFPIα and that binding of the C4BP β-chain blocks this function.
Introduction
Protein S is an essential cofactor for both the tissue factor pathway inhibitor (TFPI) and activated protein C (APC) pathways.1 In this way, protein S regulates both the initiation and propagation phases of coagulation. Consequently, human protein S deficiencies are associated with increased thrombotic risk.2 Protein S is not only present in plasma but also in platelets where it is released from α-granules following activation. Calzavarini et al recently showed that platelet protein S specifically limits venous thrombosis.3 Moreover, complete deletion of the protein S gene (PROS1) in mice results in fatal in utero coagulopathy.4,5 Protein S has an N-terminal γ-carboxylated glutamic acid (Gla) domain, a thrombin sensitive region, 4 epidermal growth factor (EGF)-like domains, and a large C-terminal sex hormone-binding globulin (SHBG)-like region, consisting of the laminin G (LG) 1 and 2 domains.1 In circulation, ∼70% of protein S is bound in a high-affinity complex with complement protein C4b binding protein (C4BP).6,7 Protein S binds to the C4BP β-chain and circulates as a protein S-C4BP complex at a 1:1 molar ratio.8 Only the free proportion of protein S (∼30%) is considered to exert full anticoagulant properties.1,9,10
The initiation phase of coagulation is regulated by TFPI. There are 2 isoforms of TFPI present in humans, TFPIα and TFPIβ. The majority of TFPI is bound to the endothelium (TFPIβ), whereas TFPIα circulates in plasma or in platelet stores where it is released following platelet activation.11,12 Protein S specifically acts as a cofactor for TFPIα.10,13 TFPIα is a Kunitz-type protease inhibitor with an acidic amino (N)-terminal end followed by 3 Kunitz domains and a basic carboxy (C)-terminal tail.11,14 It inhibits tissue factor (TF)-factor-(F)VIIa-mediated generation of FXa through the formation of a large quaternary complex by interacting with FVIIa and FXa, as well as inhibiting FXa activity directly by forming a FXa-TFPIα complex. Protein S acts as a cofactor, enhancing both of these reactions.10,15 The TFPIα-mediated FXa inhibition occurs through a 2-step reaction, first by the formation of a loose encounter complex, followed by isomerization, which results in a tight binding enzyme-inhibitor complex.11 Given its low plasma concentration (0.25-0.5 nM),11,16,17 TFPIα requires protein S to exhibit its full inhibitory potential whereby protein S stimulates the formation of the FXa-TFPIα encounter complex by reducing the Ki 4- to 10-fold in the presence of anionic phospholipids.10,18,19 The TFPI cofactor function of protein S is dependent on a direct TFPIα-protein S interaction involving the TFPIα Kunitz domain 3 and the protein S LG1 domain.18,20,21 In a previous study, we provided evidence that the protein S interaction site in the TFPIα Kunitz domain 3 includes amino acid residues Asp194, Arg199, Glu226, Glu234, and Arg237, with Arg199 and Glu226 being most important.18 Following the identification of the importance of protein S LG1 for its TFPI cofactor function, we also created composite protein S variants spanning LG1 and LG2 to further locate the TFPIα interaction site.22 Protein S variant SHBG2 with amino acid substitutions in both LG domains (E612A, I614A, F265A, V393A, and H453A) and SHBG3 with only LG1 amino acid substitutions (K317A, I330A, V336A, and D365A) showed mild to moderate reductions in TFPI cofactor function and interaction with TFPIα. The relatively mild effects that these substitutions had on the TFPI pathway suggested that the most important LG1 amino acid residues for this particular function of protein S were as yet unidentified.22
Recently, the procofactor FV19,23 and the splice variant of FV identified in individuals with east Texas bleeding disorder,24,25 FV-short, have been identified as synergistic cofactors, together with protein S, for TFPIα. The enhancement of TFPIα by FV/FV-short is dependent on the protein S SHBG-like region,19,26 which is of particular interest since this region is involved in the interaction with TFPIα as well as FV.21,22,27 However, how important the TFPIα-protein S interaction is for the synergistic enhancement by FV/FV-short is not yet known.
In addition to the TFPIα and proposed FV interaction sites, the protein S SHBG-like region also contains the C4BP interaction site. The C4BP-protein S binding likely involves extended interactions, known to include both LG domains of protein S and complement control protein (CCP) domains 1 to 2 of the C4BP β-chain.28-31 The proposed interaction between TFPIα and the protein S LG1 domain may explain why free protein S interacts with TFPIα in circulation whereas C4BP-bound protein S does not,32 resulting in reduced TFPI cofactor function of C4BP-bound protein S.10,21
In this present study, we sought to identify conclusively amino acid residues in the protein S LG1 domain, which are essential for its TFPIα enhancement/binding and their importance for the synergistic TFPI cofactor function of protein S together with FV-short. We also aimed to reveal whether the same region of protein S overlaps with the C4BP interaction site.
Methods
Generation and expression of protein S and its variants
We generated 11 protein S variants. In 7 of the variants, we substituted 1 to 2 amino acids to incorporate an N-linked glycosylation motif: D253T, I358N/K360T, L379T, E396N/I398T, R404T, G418N, and Q427N/K429T. In the remaining 4 variants, we substituted 4 to 6 surface-exposed charged residues for alanine. For this, the following mutations were incorporated: E263A/E428A/K429A/K432A (LG1A); K255A/E257A/D287A/R410A/K423A/E424A (LG1B); D304A/K323A/K392A/E394A/E396A/R404A (LG1C); and E294A/R314A/K317A/E319A/K399A/D455A (LG1D). All variants were created through site-directed mutagenesis, stably expressed in HEK293 cells and either concentrated in conditioned media or purified and quantified as previously described.21,33-35 See supplemental Methods section in supplemental Information for details.
TFPIα, FV-short, C4BP, and the isolated C4BP β-chain
TFPIα with an N-terminal 6xHis-tag (pcDNA3.1) was transiently expressed in mammalian HEK293T cells (ATCC).18 TFPIα was purified using nickel purification followed by a Heparin HiTrap HP column and was estimated by ELISA18 to be 80% to 95% full-length.
An expression vector for FV-short (east Texas bleeding disorder variant) was generated by using the full-length FV vector (pED) as a template according to a previously described method.24 FV-short was stably expressed in BHK cells, purified, and quantified as previously described.19,36,37
β-chain containing (and protein S-free) C4BP was purified from pooled fresh frozen citrated human plasma.38-40
CCP1-2 of the C4BP β-chain (Ser1-Ser119) with a C-terminal 6xHis-tag was expressed in Drosophila Schneider 2 insect cells and purified using a Ni2+-charged HisTrap Chelating HP column followed by Superdex 200 Increase 10/300 GL.
See supplemental Methods section in supplemental Information for further details on TFPIα, FV-short, and C4BP CCP1-2 vector generation, expression, and purification.
Phospholipid vesicle preparation
The phospholipids (Avanti Polar Lipids) 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-Dioleoyl-sn-glycero-3-phosphoserine (DOPS), and 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were mixed in a molar ratio of 60:20:20 and extruded as previously described.33
APC and TFPIα cofactor function in thrombin-generation assays determined by calibrated automated thrombography (CAT)
CAT was performed in protein S-depleted plasma (Enzyme Research Laboratories).18,21,33,35 Protein S (0-100 nM) was added to plasma (80 µL/well), together with 50 μM phospholipid vesicles (20:20:60; DOPS/DOPE/DOPC), and 16.6 mM CaCl2 in the presence or absence of 9 or 10 nM of APC (Haematologic Technologies Inc.) or 1 nM TFPIα. For the experiments performed in the presence of C4BP or C4BP β-chain CCP1-2, protein S was preincubated with plasma-purified C4BP or β-chain CCP1-2 at a 1:2 molar ratio for 20 minutes, which has been determined to result in fully saturated protein S in gel filtration experiments (C4BP and C4BP β-chain CCP1-2) and immunoprecipitation (C4BP β-chain CCP1-2) (data not shown). Thrombin generation was initiated by 1 pM tissue factor (Dade Innovin) in a total volume of 120 µL. Thrombin generation was monitored using 0.42 mM Z-Gly-Gly-Arg-AMC (Bachem). To inhibit contact activation, corn trypsin inhibitor (Enzyme Research Laboratories) was added (65 µg/mL plasma). All given concentrations are final.
FXa inhibition assay
The inhibition of FXa (0.5 nM; Enzyme Research Laboratories) activity by TFPIα was monitored by the cleavage of the chromogenic substrate S-2765 (200 μM; Chromogenix). For TFPIα enhancement by protein S alone, this was measured in the presence or absence of 2.5 nM TFPIα and protein S (0-160 nM), 25 μM phospholipid vesicles, and 5 mM CaCl2, as described previously.18,20,21 When measuring the TFPIα enhancement by protein S together with FV-short, the same experiments were performed in the presence or absence of 0.5 nM TFPIα, 5 nM protein S, and 0 to 4 nM FV-short. To derive the concentration of protein S or FV-short required to reach 50% of the maximal potentiation of TFPIα-mediated inhibition of FXa (EC50), the initial velocity (V0) of the S-2765 cleavage was plotted against the protein S or FV-short concentration, respectively. The V0 was determined using nonlinear regression as previously described.10,20 The EC50 was determined by a 1-phase exponential decay nonlinear curve fit.18,21
Coimmunoprecipitation of TFPIα-protein S
Coimmunoprecipitation experiments were used to investigate the TFPIα-protein S interaction. Briefly, 50 nM TFPIα was preincubated with 40 nM protein S in concentrated conditioned media (150 μL) for 30 minutes to allow interaction. Following incubation, TFPIα, together with any bound protein S, was pulled down from solution using tosylactivated magnetic beads coated with polyclonal anti-human TFPI antibodies (0.2 mg beads for 150 μL sample). Bound proteins were eluted and analyzed by western blotting. See supplemental Materials for further details.
Protein S-C4BP/C4BP β-chain binding
Analysis of protein S binding to C4BP or C4BP β-chain CCP1-2 was performed using plate-binding assays essentially as described.22,41 Protein S (0-20 nM) was incubated on plates coated with C4BP, followed by detection with anti-protein S antibodies. Alternatively, protein S was coated on the plate, followed by incubation with increasing concentrations of C4BP β-chain CCP1-2 and detection of C4BP β-chain CCP1-2 binding using HRP-conjugated rabbit anti-His antibodies. See supplemental Materials for further details.
Results
Screening for LG1 regions involved in TFPI cofactor function
To screen the surface of the protein S LG1 domain for its involvement in the TFPI cofactor function of protein S, N-linked glycosylation motifs were introduced into the protein S cDNA. Seven variants were generated and expressed, with glycan motifs introduced over the LG1 domain (Figure 1A). Of the 7 variants, 5 were successfully expressed. Variant E396N/I398T did not express, whereas I358N/K360T repeatedly precipitated during concentration, suggesting protein misfolding. Both variants were therefore excluded from further investigations. While protein S variants D253T, L379T, G418N, and Q427N/K429T exhibited a mobility shift on SDS-PAGE, consistent with the incorporation of an additional N-linked glycosylation, this was not observed in protein S R404T (supplemental Figure S1A), suggesting either no or modest influence on C4BP binding. However, as the expected mobility shifts were small, all 5 variants were included in the following analyses. Wild-type (WT) protein S and its variants were initially tested for their ability to enhance APC in plasma-based CAT screening assays. For this, 50 nM protein S in concentrated conditioned media was added to protein S-depleted plasma, supplemented with 9 nM APC. Thrombin generation was initiated with 1 pM TF and followed in real-time. No significant difference in APC enhancement was observed for any of the variants, suggesting that the mutations did not influence APC cofactor function (Figure 1B; supplemental Figure S2A). The variants were also tested for their affinities for C4BP.
APC and TFPI cofactor function of N-linked glycan protein S variants determined using CAT. (A) Spatial distribution of amino acids which were substituted to obtain additional N-linked glycosylations in the protein S SHBG-like region are highlighted in red. Naturally occurring N-linked glycolysations are highlighted in blue. The model of the SHBG-like region is adapted from Villoutreix et al.42 (B-E) The generated variants were investigated for their abilities to function as cofactors for APC (B) or TFPIα (C-E) using CAT. For this, thrombin generation was initiated by 1 pM TF in protein S-depleted plasma supplemented with 50 μM phospholipids. (B) Screening assay for thrombin generation quantified in plasma supplemented with 9 nM APC in the presence or absence of 50 nM WT or variant protein S. The relative APC enhancement by the protein S variants were determined as the reduction in thrombin peak height compared with that of WT protein S. The decrease in peak height in the presence of WT protein S compared with APC alone was set as 100%. (C) Screening assay for TFPI cofactor function of the protein S variants, using thrombin generation measured in plasma supplemented with 1 nM TFPIα in the presence or absence of 50 nM WT or variant protein S. The relative enhancement of TFPIα was calculated as that of APC above. Here, the decrease in peak height in the presence of both WT protein S and TFPIα compared with that caused by TFPIα alone was set as 100%. (D-E) Titrations of TFPI cofactor function were performed for WT protein S, protein S D253T, and Q427N/K429T (0-50 nM), and the normalized lag times (D) and peak thrombin (E) were plotted against protein S concentration. The peak height and lag time observed in the presence of TFPIα alone represents 100%. The results are presented as mean ± SEM (n = 3-6). PS, protein S.
APC and TFPI cofactor function of N-linked glycan protein S variants determined using CAT. (A) Spatial distribution of amino acids which were substituted to obtain additional N-linked glycosylations in the protein S SHBG-like region are highlighted in red. Naturally occurring N-linked glycolysations are highlighted in blue. The model of the SHBG-like region is adapted from Villoutreix et al.42 (B-E) The generated variants were investigated for their abilities to function as cofactors for APC (B) or TFPIα (C-E) using CAT. For this, thrombin generation was initiated by 1 pM TF in protein S-depleted plasma supplemented with 50 μM phospholipids. (B) Screening assay for thrombin generation quantified in plasma supplemented with 9 nM APC in the presence or absence of 50 nM WT or variant protein S. The relative APC enhancement by the protein S variants were determined as the reduction in thrombin peak height compared with that of WT protein S. The decrease in peak height in the presence of WT protein S compared with APC alone was set as 100%. (C) Screening assay for TFPI cofactor function of the protein S variants, using thrombin generation measured in plasma supplemented with 1 nM TFPIα in the presence or absence of 50 nM WT or variant protein S. The relative enhancement of TFPIα was calculated as that of APC above. Here, the decrease in peak height in the presence of both WT protein S and TFPIα compared with that caused by TFPIα alone was set as 100%. (D-E) Titrations of TFPI cofactor function were performed for WT protein S, protein S D253T, and Q427N/K429T (0-50 nM), and the normalized lag times (D) and peak thrombin (E) were plotted against protein S concentration. The peak height and lag time observed in the presence of TFPIα alone represents 100%. The results are presented as mean ± SEM (n = 3-6). PS, protein S.
While protein S L379T and G419N both showed a statistically significant reduction in C4BP binding affinity, all variants bound to C4BP with high affinity (supplemental Table S1). The ability of the protein S variants to enhance TFPIα was also tested in CAT assays. For this, 50 nM protein S in concentrated conditioned media was added to protein S-depleted plasma. The protein S depletion process also results in partial codepletion of TFPI (0.39 ± 0.04 nM vs 1.73 ± 0.18 nM in normal plasma) but more importantly, complete depletion of all TFPIα (undetectable vs 0.34 ± 0.08 nM in normal plasma).18,21 Protein S-depleted plasma was therefore supplemented with 1 nM TFPIα. While the majority of protein S variants enhanced TFPIα equally well as WT protein S, protein S variants D253T and Q427N/K429T exhibited reduced TFPIα enhancement (Figure 1C; supplemental Figure S2B). Titration of protein S D253T and Q427N/K429T revealed a severe reduction in TFPI cofactor function compared with WT protein S. This was observed through a diminished reduction in peak height, but more evidently by the reduced prolongation of the lag time before thrombin generation was initiated (Figure 1D-E).
Identification of protein S LG1 residues important for TFPI cofactor function
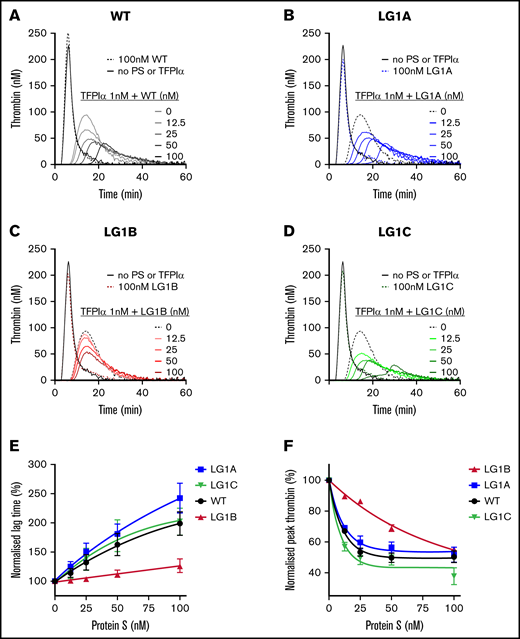
We next generated 4 composite variants in which clusters of surface-exposed charged residues (4-6 amino acids) were substituted to alanine. Three of these, LG1A, LG1B, and LG1C, spanned regions in close proximity to the inserted N-linked glycan attachment sites in protein S D253T and Q427N/K429T. The fourth variant, LG1D, spanned regions around the previously described protein S SHBG3 (K317A, I330A, V336A, and D365A), a variant suggested to be close to the TFPIα interaction site (Figure 2).22 Of the 4 composite protein S variants, LG1D was not expressed and was therefore excluded from further studies. The remaining 3 variants, LG1A, LG1B, and LG1C, were expressed at similar levels to WT protein S and purified (supplemental Figure S1B). The 3 variants were investigated for their TFPI cofactor function in CAT assays, alongside WT protein S. Protein S was titrated (0-100 nM) in the presence of 1 nM TFPIα. WT protein S enhanced TFPI function as evidenced primarily by prolongation of the lag time but also by reduction in the peak thrombin (Figure 3A,E).22 While both LG1A and LG1C variants enhanced TFPIα-mediated inhibition of thrombin generation similar to WT protein S, protein S LG1B exhibited markedly reduced TFPIα enhancement (Figure 3).
Spatial distribution of amino acid substitutions in the protein S LG1 domain. Charged residues in the protein S LG1 domain were substituted to alanine. Four composite protein S variants were generated, each containing 4 to 6 amino acid substitutions chosen based on their spatial proximity to the sites where insertion of N-linked glycans led to a reduction in TFPI cofactor function or to amino acid residues previously suggested to be functionally important for the TFPIα enhancement. The model of the SHBG-like region is adapted from Villoutreix et al.42
Spatial distribution of amino acid substitutions in the protein S LG1 domain. Charged residues in the protein S LG1 domain were substituted to alanine. Four composite protein S variants were generated, each containing 4 to 6 amino acid substitutions chosen based on their spatial proximity to the sites where insertion of N-linked glycans led to a reduction in TFPI cofactor function or to amino acid residues previously suggested to be functionally important for the TFPIα enhancement. The model of the SHBG-like region is adapted from Villoutreix et al.42
Enhancement of the TFPIα-mediated inhibition of thrombin generation by WT protein S and alanine composite variants. Thrombin generation was measured in protein S-depleted plasma supplemented with 50 µM phospholipids and 1 pM TF in the presence and absence of 1 nM TFPIα, and in the presence and absence of titrated (0-100 nM) WT protein S (A), protein S LG1A (B), LG1B (C), and LG1C (D). Representative experiments are shown (n = 3-4). (E-F) Normalized lag time (E) or peak thrombin (F) were plotted against protein S concentration. The lag time and peak height in the presence of TFPIα alone represent 100%. Results are expressed as mean ± SD (n = 3-4). PS, protein S.
Enhancement of the TFPIα-mediated inhibition of thrombin generation by WT protein S and alanine composite variants. Thrombin generation was measured in protein S-depleted plasma supplemented with 50 µM phospholipids and 1 pM TF in the presence and absence of 1 nM TFPIα, and in the presence and absence of titrated (0-100 nM) WT protein S (A), protein S LG1A (B), LG1B (C), and LG1C (D). Representative experiments are shown (n = 3-4). (E-F) Normalized lag time (E) or peak thrombin (F) were plotted against protein S concentration. The lag time and peak height in the presence of TFPIα alone represent 100%. Results are expressed as mean ± SD (n = 3-4). PS, protein S.
Protein S variant LG1B residues are important for TFPIα interaction
The interaction between WT protein S and its variants with TFPIα was investigated in solution using coimmunoprecipitation. For this, 40 nM protein S was preincubated with 50 nM TFPIα followed by immunoprecipitation using magnetic beads coated with anti-TFPI K1-K2 antibodies that do not interfere with TFPIα-protein S. Bound proteins were eluted and analyzed by western blotting. WT protein S was coimmunoprecipitated together with TFPIα (supplemental Figure S3). In contrast, little WT protein S was detected in the eluates from beads incubated with WT protein S alone, in the absence of TFPIα, suggesting that the binding was specific for TFPIα. Variant LG1C bound equally well to TFPIα as WT protein S (120% binding compared with WT) whereas a reduced binding was observed for LG1A (33% binding) and very little TFPIα binding was seen for protein S LG1B (12% binding), suggesting severely reduced binding affinity.
Protein S variant LG1B does not enhance TFPIα-mediated inhibition of FXa
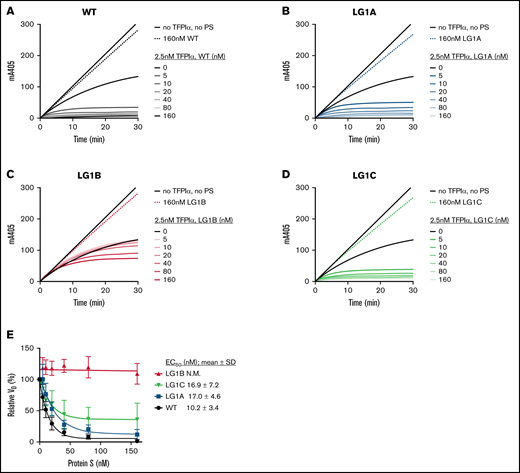
The TFPI cofactor functions of WT protein S and its variants were investigated in the direct inhibition of FXa. In contrast to direct binding assays, which have been previously used to establish the dissociation constant of the TFPIα-interaction to ∼1 μM, functional assays, such as the direct FXa inhibition assays, have suggested a much higher affinity.18,20,21,32 For this reason, a real-time, pure-component FXa inhibition assay was employed to further evaluate the importance of the substituted protein S residues as part of a functional interaction site. Progress curves of FXa inhibition by 2.5 nM TFPIα and increasing concentrations of protein S (0-160 nM) showed that protein S LG1A and LG1C functioned equally well as TFPIα cofactors as WT protein S (Figure 4A,B,D). However, and in agreement with the plasma-based CAT assays and the reduced affinity observed in the coimmunoprecipitation experiments (Figure 3; supplemental Figure S3), protein S LG1B was a poor TFPIα cofactor and showed a limited effect on the TFPIα-mediated inhibition of FXa activity (Figure 4C). The initial rates of S-2765 hydrolysis by FXa in the presence of TFPIα and increasing concentrations of protein S are plotted in Figure 4E, and the EC50 values have been derived. The EC50 for TFPIα-mediated enhancement by protein S LG1C (16.9 ± 7.2 nM) was not statistically significantly different from that of WT protein S (10.2 ± 3.4 nM). Neither was that of LG1A (17.0 ± 4.6 nM), which was somewhat surprising considering the observed reduction in TFPIα-binding (supplemental Figure S3). In contrast, the EC50 for TFPIα enhancement by LG1B could not be quantitated due to its very low cofactor function, which agreed with the almost complete lack of interaction observed in the coimmunoprecipitation assays.
Enhancement of TFPIα in the inhibition of FXa by WT protein S and protein S alanine composite variants. FXa activity (0.5 nM) was followed in real-time through cleavage of S-2765 (200 µM) at 405 mm in the presence of 25 μM phospholipids, presence or absence of TFPIα (2.5 nM), and increasing concentrations (0-160 nM) of WT protein S (A), LG1A (B), LG1B (C), or LG1C (D). Results from representative experiments are shown (n = 3-4). (E) The initial velocity (V0) was calculated for each curve and plotted against protein S concentration. Results are given as mean ± SD and are expressed as a percentage of the V0 for TFPIα alone (n = 3-4). The EC50 values of TFPIα enhancement by WT protein S and its variants were derived and are presented as part of the figure. N.M., not measurable; PS, protein S.
Enhancement of TFPIα in the inhibition of FXa by WT protein S and protein S alanine composite variants. FXa activity (0.5 nM) was followed in real-time through cleavage of S-2765 (200 µM) at 405 mm in the presence of 25 μM phospholipids, presence or absence of TFPIα (2.5 nM), and increasing concentrations (0-160 nM) of WT protein S (A), LG1A (B), LG1B (C), or LG1C (D). Results from representative experiments are shown (n = 3-4). (E) The initial velocity (V0) was calculated for each curve and plotted against protein S concentration. Results are given as mean ± SD and are expressed as a percentage of the V0 for TFPIα alone (n = 3-4). The EC50 values of TFPIα enhancement by WT protein S and its variants were derived and are presented as part of the figure. N.M., not measurable; PS, protein S.
Protein S variant LG1B is a weak synergistic TFPI cofactor together with FV-short
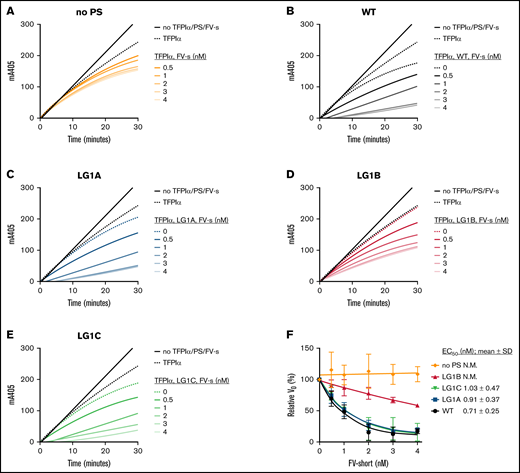
The enhancement of TFPIα-mediated inhibition of FXa was also investigated in the presence of FV-short. Progress curves of FXa inhibition by 0.5 nM TFPIα were followed in the presence or absence of 5 nM protein S and increasing concentrations of FV-short (0-4 nM). As expected from previous studies, FV-short had little effect on TFPIα-mediated inhibition of FXa in the absence of protein S (Figure 5A). WT protein S enhanced TFPIα in the absence of FV-short. However, the enhancement was appreciably enhanced by FV-short (Figure 5A-B). Similar synergistic enhancement was also observed for protein S LG1A and LG1C (Figure 5C,E). Protein S LG1B showed no enhancement in the absence of FV-short, in agreement with the data presented in Figure 4. However, a moderate enhancement was observed by LG1B together with FV-short, suggesting that the reduced TFPIα-LG1B affinity also reduced the variant’s ability to function as a synergistic TFPI cofactor together with FV-short, but did not abolish it completely (Figure 5D). The initial rates of S-2765 hydrolysis by FXa in the presence of TFPIα and WT protein S and its variants were plotted at increasing concentrations of FV-short, and the EC50 values were derived (Figure 5E). The EC50 for TFPIα-mediated synergistic enhancement by protein S LG1A (0.91 ± 0.37 nM) and LG1C (1.03 ± 0.47 nM) together with FV-short were not statistically significantly different from that of WT protein S (0.71 ± 0.25 nM). While some synergistic enhancement was detected by LG1B, the EC50 was too high to be reliably quantified.
Enhancement of TFPIα in the inhibition of FXa by increasing concentrations of FV-short in the presence or absence of WT protein S and protein S alanine composite variants. FXa activity (0.5 nM) was followed in real-time through cleavage of S-2765 (200 µM) at 405 mm in the presence of 25 μM phospholipids, TFPIα (0.5 nM) and the absence (A) or presence of 5 nM of WT protein S (B), LG1A (C), LG1B (D), or LG1C (E) at increasing concentrations of (F) FV-short (0-4 nM). Results from representative experiments are shown (n = 3). (E) The initial velocity (V0) was calculated for each curve and plotted against FV-short concentration. Results are given as mean ± SD and are expressed as percentage of the V0 for TFPIα either alone or together with WT protein S or its variants (n = 3). The EC50 values of TFPIα enhancement by FV-short and WT protein S and its variants were derived and are presented as part of the figure. FV-s, FV-short; N.M., not measurable; PS, protein S.
Enhancement of TFPIα in the inhibition of FXa by increasing concentrations of FV-short in the presence or absence of WT protein S and protein S alanine composite variants. FXa activity (0.5 nM) was followed in real-time through cleavage of S-2765 (200 µM) at 405 mm in the presence of 25 μM phospholipids, TFPIα (0.5 nM) and the absence (A) or presence of 5 nM of WT protein S (B), LG1A (C), LG1B (D), or LG1C (E) at increasing concentrations of (F) FV-short (0-4 nM). Results from representative experiments are shown (n = 3). (E) The initial velocity (V0) was calculated for each curve and plotted against FV-short concentration. Results are given as mean ± SD and are expressed as percentage of the V0 for TFPIα either alone or together with WT protein S or its variants (n = 3). The EC50 values of TFPIα enhancement by FV-short and WT protein S and its variants were derived and are presented as part of the figure. FV-s, FV-short; N.M., not measurable; PS, protein S.
Amino acid residues important for TFPI cofactor function are not important for APC cofactor function but may be involved in C4BP binding
The protein S variants were also assessed for their APC cofactor function and affinity for C4BP. For this, increasing concentrations of protein S (0-50 nM) were added to protein S-depleted plasma supplemented with 10 nM APC. Protein S LG1C showed slightly reduced APC cofactor function compared with WT protein S (supplemental Figure S4A,D). In contrast, protein S LG1A and LG1B enhanced APC-mediated reduction of thrombin generation equally well as WT protein S (supplemental Figure S4). Some of the amino acid residues substituted in protein S LG1C were in very close proximity to the LG2 domain, previously shown to be most important for APC enhancement, which could be a cause of the moderately reduced APC cofactor function observed for this variant.
All variants showed statistically significant differences in binding affinities compared with WT protein S (Table 1), with their binding to C4BP still being of high affinity. While these results suggest that the C4BP and TFPIα interaction sites are distinct, it cannot be excluded that they may be in close proximity to each other or potentially partially overlapping.
Evaluation of binding of WT protein S and composite protein S variants to C4BP
| Protein S variant . | Kd (nM) . |
|---|---|
| WT | 0.67 ± 0.18 |
| LG1A | 1.21 ± 0.31** |
| LG1B | 1.13 ± 0.37* |
| LG1C | 1.40 ± 0.35** |
| Protein S variant . | Kd (nM) . |
|---|---|
| WT | 0.67 ± 0.18 |
| LG1A | 1.21 ± 0.31** |
| LG1B | 1.13 ± 0.37* |
| LG1C | 1.40 ± 0.35** |
Serial dilutions (0-20 nM) of protein S were incubated with 5 µg/mL plate-bound C4BP. Bound protein S was detected with monoclonal antiprotein S antibodies. The affinity of the interaction was derived using a 1-site binding equation. The average of Kd values is shown as mean ± SD (n = 6).
P < .05, **P < .01; evaluated by Mann Whitney test.
C4BP β-chain-binding blocks protein S residues required for efficient TFPIα enhancement
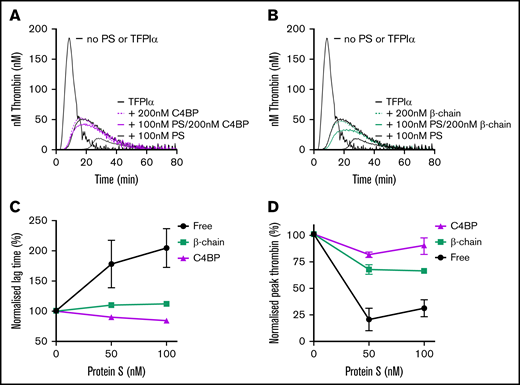
To further investigate a potential overlap of the TFPIα and C4BP interaction sites in protein S, we expressed and purified the recombinant C4BP β-chain CCP1-2 (supplemental Figure S5), which is much smaller (25 kDa) than the entire C4BP molecule but contains the full interaction site.28-31 C4BP β-chain CCP1-2 binding to protein S was similar to that of C4BP (KD of 0.32 ± 0.09 nM vs 0.67 ± 0.18 nM). The functional consequences of protein S binding to C4BP β-chain CCP1-2 were compared with that of C4BP-bound and free protein S in CAT assays. For this, protein S was incubated with a 2-fold molar excess of C4BP or β-chain CCP1-2 for 30 minutes to ensure full saturation of protein S. Protein S-depleted plasma was reconstituted with 1 nM TFPIα in the presence or absence of 50 or 100 nM protein S, or protein S bound to either C4BP or β-chain CCP1-2. Free protein S efficiently enhanced TFPIα in the inhibition of thrombin generation. C4BP-bound protein S exhibited very little TFPI cofactor function (Figure 6A,C,D). Similarly, the isolated C4BP β-chain CCP1-2 also almost completely abolished the TFPI cofactor function of protein S (Figure 6B-D). These results suggest that the β-chain binding site may partially overlap or lie in close proximity to that of TFPIα.
Enhancement of the TFPIα-mediated inhibition of thrombin generation by free protein S compared with that of protein S bound to C4BP or the isolated C4BP β-chain. Thrombin generation was measured in protein S-depleted plasma supplemented with 50 µM phospholipids and 1 pM TF in the presence and absence of 1 nM TFPIα, and in the presence and absence of protein S (50 or 100 nM) which was either free (A-B) or prebound to saturating (100 or 200 nM) concentrations of the entire C4BP molecule (A) or the isolated C4BP β-chain (B). Representative experiments are shown (n = 4-5). (C-D) Normalized lag time (C) or peak thrombin (D) were plotted against protein S concentration. The lag time and peak height in the presence of TFPIα alone represent 100%. Results are expressed as mean ± SD (n = 4-5). PS, protein S.
Enhancement of the TFPIα-mediated inhibition of thrombin generation by free protein S compared with that of protein S bound to C4BP or the isolated C4BP β-chain. Thrombin generation was measured in protein S-depleted plasma supplemented with 50 µM phospholipids and 1 pM TF in the presence and absence of 1 nM TFPIα, and in the presence and absence of protein S (50 or 100 nM) which was either free (A-B) or prebound to saturating (100 or 200 nM) concentrations of the entire C4BP molecule (A) or the isolated C4BP β-chain (B). Representative experiments are shown (n = 4-5). (C-D) Normalized lag time (C) or peak thrombin (D) were plotted against protein S concentration. The lag time and peak height in the presence of TFPIα alone represent 100%. Results are expressed as mean ± SD (n = 4-5). PS, protein S.
Discussion
How protein S functions as a cofactor in the TFPI pathway remains poorly understood. Certainly, protein S has to interact with TFPIα to enhance its anticoagulant functions.18,20 This interaction involves the protein S LG1 domain and Glu226 and Arg199 in the TFPIα Kunitz domain 3.18,20-22 However, TFPIα Kunitz domain 3 binding sites in protein S LG1 have not been fully elucidated. In a previous study, we identified a cluster of amino acids on protein S which, when substituted to alanine, reduced the affinity of protein S toward TFPIα, resulting in a moderate reduction in TFPI cofactor function.22 In the present study, we screened large surface areas in the entire LG1 domain for their functional importance for the protein S TFPI cofactor function by inserting N-linked glycan attachment sites that might block larger regions of the surface of the LG1 domain. We found that inserting N-linked glycan attachment sites at protein S LG1 amino acid positions 253 and 427 reduced their TFPI cofactor function. These results suggested that areas surrounding these amino acids are functionally important for the TFPI cofactor function of protein S. Both sites are in close proximity to each other in the protein S LG1 domain, further suggesting the importance of this region. We then substituted proximal surface-exposed charged residues to alanine in clusters. One variant, K255A/E257A/D287A/R410A/K423A/E424A (LG1B), showed almost no TFPI cofactor function either in plasma-based thrombin generation assays or in pure-component FXa inhibition assays. These residues are adjacent to the 2 N-linked glycosylation site variants, N253T and Q427N/K429T, supporting the initial screening results that this particular region of LG1 is important for the TFPI cofactor function of protein S. Although the SHBG-like region has also been suggested to be involved in the APC cofactor function of protein S and C4BP binding, substitution of these amino acids did not influence the ability of protein S to enhance APC with very little effect on C4BP binding-affinity. Variant LG1B also bound appreciably less efficiently to TFPIα in coimmunoprecipitation assays, suggesting that the reason for the reduced TFPI cofactor function was due to a reduction in direct-binding affinity. A moderate reduction in binding was also observed for protein S LG1A. However, in contrast to LG1B, this did not diminish its cofactor function in any of the functional assays. Protein S LG1B was also a poor synergistic cofactor, together with FV-short. These results suggest that while some synergistic TFPIα enhancement can occur by protein S and FV-short also if the TFPIα-protein S interaction is impeded, an optimal interaction is required for a fully functional TFPI pathway. These results could also explain why some limited TFPI cofactor functions of protein S LG1B remained in our plasma-based assays since FV species that can act as a synergistic cofactor for TFPIα, together with protein S, are very likely present there.
It is already established that C4BP-binding essentially abolishes the TFPI cofactor function of protein S.10,21 However, while both TFPIα and C4BP interact with the protein S LG1 domain, the reason for the reduction in TFPI cofactor function of C4BP-bound protein S was not known. Even though protein S only interacts with the C4BP β-chain,29-31 the 570kDa C4BP may sterically block access to the protein S LG domains rather than compete for the same binding site as TFPIα. Our protein S variants did not enable us to clarify this issue. Instead, using purified recombinant C4BP β-chain CCP1-2, we demonstrated that this, like C4BP, severely diminishes TFPI cofactor function of protein S. This finding was somewhat surprising considering the high-affinity binding observed for protein S LG1B to C4BP. If the C4BP interaction site overlaps with amino acid residues that are functionally important for the protein S TFPI cofactor function, one may have expected a larger reduction in C4BP-binding affinity. However, in contrast to the affinity of protein S toward TFPIα (determined as 1 μM in direct binding assays or ∼10 to 20 nM in functional assays),20-22,32 the binding of protein S toward C4BP is of much higher affinity (determined here as 0.73 nM). The protein S-C4BP interaction has also been shown to span both protein S LG1 and LG2. Overall, our results suggest TFPIα and C4BP interactions are distinct but in close proximity to each other.
To summarize, the results presented in this study show that protein S Lys255, Glu257, Asp287, Arg410, Lys423, and Glu424 are important for protein S-mediated enhancement of TFPIα, both in the presence and absence of FV-short. They also show that binding of the C4BP β-chain blocks this function of protein S.
Acknowledgments
This research was funded by the British Heart Foundation (PG/14/37/30855, RG/18/3/33405, PG/20/13/34994, and PG/18/15/33566).
Authorship
Contribution: A.T.-O., M.G., A.P., and J.A. designed the research, performed experiments, analyzed the results, and wrote the paper; D.J., R.K., P.B.F., and S.S. performed the experiments, analyzed the results, and revised the manuscript; and J.T.B.C. and D.A.L. designed the research and revised the manuscript.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Josefin Ahnström, Centre for Haematology, Imperial College London, Fifth Floor Commonwealth Building, Hammersmith Hospital Campus, Du Cane Road, London W12 0NN, United Kingdom; e-mail: j.ahnstrom@imperial.ac.uk.
References
Author notes
For data sharing, please contact the corresponding author at j.ahnstrom@imperial.ac.uk.
The full-text version of this article contains a data supplement.