TO THE EDITOR:
Next-generation sequencing (NGS) has demonstrated the existence of recurrent activating mutations in STAT3 and STAT5B in ∼40% to 50% of patients with T-cell large granular lymphocytic leukemia (T-LGLL) and 20% to 30% of patients with natural killer (NK) cell granular lymphocytic leukemia (NK-LGLL; also known as chronic lymphoproliferative disorder of NK cells).1-3STAT3/STAT5B mutations alone are not specific to T- or NK-LGLL, because they have frequently been found in other lymphoid neoplasms (LNs), as well as in myeloid neoplasms (MNs), aplastic anemia, and other autoimmune disorders and rarely in asymptomatic individuals with clonal hematopoiesis of indeterminate potential.1,4-6 Compounding the issue of specificity is that clonal large granular lymphocyte (LGL) expansions are frequently observed in MNs, and frequent STAT3/STAT5B mutations have been reported in MNs with associated LGL expansion.4,7,8 LGLLs have rarely been diagnosed concomitantly in patients with otherwise typical MNs.9,10
Based on morphology, flow cytometry, and T-cell clonality studies alone, it may be challenging to classify a marrow process with both LGL expansion11-14 and morphologic dysplasia7 (supplemental Figure 1) as an LGLL vs an MN or both.7 To present further challenges, both manifest with varying degrees of cytopenias and heterogenous lymphocyte counts and are frequently found in older people. Genetic profiling using NGS supports an accurate diagnosis in patients with diagnostic challenges and is an important area of investigation because patients with cytopenia with different underlying causes require different therapies.10
In our multi-institutional study, we examined how a targeted myeloid NGS panel can distinguish T- or NK-LGLLs from MNs. We retrospectively identified 118 patients with hematologic neoplasms (66 patients with LNs, 50 with MNs, and 2 with concomitant LGLL and myelodysplastic syndrome [MDS]; Figure 1A), all with pathogenic or likely pathogenic STAT3/STAT5B variants. We also identified 18 patients with MDS who had clonal LGL expansions8,15 in the absence of STAT3/STAT5B mutations (Figure 1A) among an unselected cohort of 6690 patients who underwent NGS testing (supplemental Tables 1 and 2). We then analyzed the demographic and genomic profiles, clonal metrics of STAT3/STAT5B variants, T-cell clonality, and flow cytometric studies (Figure 1A; supplemental Table 3). Details regarding materials and methods are available in the supplemental Materials.
Comprehensive genetic profiles, hotspots, and VAF distribution of STAT3 and STAT5B variants in LNs and MNs. (A) LNs are displayed on the left (light blue, top row) and MNs on the right (light orange, top row). The World Health Organization (WHO) entities in each group are listed in the second row. The concomitant variants were grouped into the following 6 categories on the basis of their gene function: Epigenetic: epigenetic regulators, genes involving DNA methylation or histone acetylation and deacetylation (light green); SFs: RNA splicing factors (purple); TFs: transcription factors (orange); Signaling: molecules in tyrosine kinase pathway or RAS/MAPK pathways (pink); C: cohesins (light purple); and Others: genes with functions beyond the above categories (various colors). Each column represents 1 patient, each bar represents 1 variant, and split bars indicated 2 or more variants in the same gene. (B) LNs showed predominantly STAT3 variants compared with (C) STAT5B variants (P = .0003), and variants seemed to be concentrated in the SH2 domain of both genes. Among STAT3 variants, in LNs, the D661 variants (including both D661V and D661Y) were most prevalent followed by Y640F. In contrast, in MNs, D661Y, not D661V, was the most common hotspot mutation followed by S614R and Y640F. (C) The STAT5BN642H variant was particularly represented among MNs and only rarely detected in LNs. (D) The distribution of VAFs among LGLLs peaked at a median of 8.8% (solid line; range, 1.4%-48.6%). (E) The VAF distribution in MNs seemed wider with a median VAF of 12.0% (solid line; range, 1.1%-65.2%; [∗] P = .01 by unpaired Student t test) and a significant second population concentrated around 48% to 50% (P = .006 by χ2 test for trend). The dashed lines represent the 25th and 75th percentiles. The relative density of VAF distribution is displayed as previously described.21 AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; MPN, myeloproliferative neoplasm; Neg, negative; NI, no information; NL, normal; NOS, not otherwise specified; SH2, Src homology 2 domain; TAD, topologically associating domain.
Comprehensive genetic profiles, hotspots, and VAF distribution of STAT3 and STAT5B variants in LNs and MNs. (A) LNs are displayed on the left (light blue, top row) and MNs on the right (light orange, top row). The World Health Organization (WHO) entities in each group are listed in the second row. The concomitant variants were grouped into the following 6 categories on the basis of their gene function: Epigenetic: epigenetic regulators, genes involving DNA methylation or histone acetylation and deacetylation (light green); SFs: RNA splicing factors (purple); TFs: transcription factors (orange); Signaling: molecules in tyrosine kinase pathway or RAS/MAPK pathways (pink); C: cohesins (light purple); and Others: genes with functions beyond the above categories (various colors). Each column represents 1 patient, each bar represents 1 variant, and split bars indicated 2 or more variants in the same gene. (B) LNs showed predominantly STAT3 variants compared with (C) STAT5B variants (P = .0003), and variants seemed to be concentrated in the SH2 domain of both genes. Among STAT3 variants, in LNs, the D661 variants (including both D661V and D661Y) were most prevalent followed by Y640F. In contrast, in MNs, D661Y, not D661V, was the most common hotspot mutation followed by S614R and Y640F. (C) The STAT5BN642H variant was particularly represented among MNs and only rarely detected in LNs. (D) The distribution of VAFs among LGLLs peaked at a median of 8.8% (solid line; range, 1.4%-48.6%). (E) The VAF distribution in MNs seemed wider with a median VAF of 12.0% (solid line; range, 1.1%-65.2%; [∗] P = .01 by unpaired Student t test) and a significant second population concentrated around 48% to 50% (P = .006 by χ2 test for trend). The dashed lines represent the 25th and 75th percentiles. The relative density of VAF distribution is displayed as previously described.21 AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; MPN, myeloproliferative neoplasm; Neg, negative; NI, no information; NL, normal; NOS, not otherwise specified; SH2, Src homology 2 domain; TAD, topologically associating domain.
There was a significant predominance of males in T-LGLLs (70%) and NK-LGLLs (100%) as well as in all other LNs (91%), whereas that predominance was not observed to the same extent in MNs (52% males; supplemental Table 4). The median age at the time of LGLL diagnosis was 71 to 73 years (range, 19-89 years), which was similar to the ages of patients with other lymphoproliferative disorders (LPDs) or MNs (supplemental Table 4).
LNs showed predominantly STAT3 variants compared with STAT5B variants (92% in LNs vs 65% in MNs; P = .0003; Figure 1B-C), and variants seemed to be concentrated in the Src homology 2 (SH2) domain of both genes. Among STAT3 variants, the D661 and Y640F variants were more prevalent in LNs, and S614R and G618R variants were common in both LNs and MNs, similar to data reported in the literature2,4 (Figure 1B-C). p.N642H was the most common variant in STAT5B in MNs, which is comparable to data reported in the literature.5,16STAT5B variants were rare in LGLLs in contrast to MNs in this retrospective study (4% in STAT3/STAT5B-mutant LGLLs vs 34% in STAT3/STAT5B-mutant MNs; P < .0001; Figure 1A), similar to the rates in previous case series.2,4,17-20 The median variant allele frequency (VAF) of STAT3/STAT5B variants21 was 8.8 (range, 1.4-48.6) among LGLLs and 12.0 in MNs (range, 1.1-65.2; P = .01; Figure1D-E). There seemed to be a bimodal and broader distribution in MNs with 2 predominant populations of around 10% or between 40% and 50% (Figure 1E), whereas most VAFs were between 5% and 18% in LGLLs (dashed lines in Figure 1D).
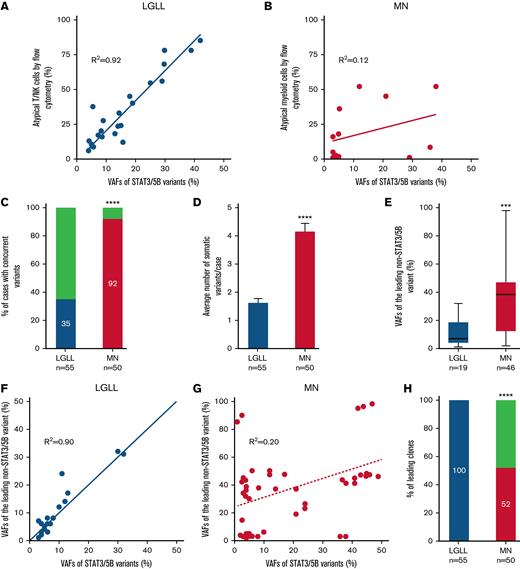
Furthermore, there was a robust correlation (Figure 2A-B) between the VAFs of STAT3/STAT5B variants and the percentage of aberrant T- or NK-LGLLs detected by flow cytometry (Figure 2A), whereas such a correlation did not exist in MNs (Figure 2B). These findings suggested that STAT3/STAT5B variants were the founder clones in LGLLs that were driving neoplastic cell proliferation. Among LGLLs, concomitant mutations were uncommon (35% in LGLLs vs 92% in MNs; P < .0001; Figure 2C) and most occurred in genes involved in epigenetic regulation (light green in Figure 1A). In contrast, MNs showed more complex genetic profiles (Figure 1A; supplemental Table 3) demonstrated by a greater number of variants (1.7 variants per patient in those with LGLLs vs 4.2 per patient in those with MNs; P < .0001; Figure 2D), greater diversity of concomitant variants (Figure 1A), and greater VAFs of the leading non-STAT3/STAT5B variants (Figure 2E) compared with LGLLs. In LGLLs, a strong correlation arose between the VAFs of STAT3/STAT5B variants and those of the leading non- STAT3/STAT5B variants or clones (R2= 0.90; Figure 2F), suggesting that STAT3/STAT5B variants were the leading clones in all LGLLs (100% in LGLLs vs 52% in MNs; P < .0001; Figure 4F). In MNs, 48% of STAT3/STAT5B variants seemed to be subclones (Figure 2G-H). The complex genetic profiles in MNs were also reflected by the otherwise similar genetic characteristics in MNs with LGL expansions detected by flow cytometry in the absence of STAT3/STAT5B mutations (18 right-most columns in Figure 1A). These findings may be the result of the myeloid-directed NGS panel used in our study and should not be interpreted to mean that LGLLs generally harbor fewer total mutations.22
Correlation of STAT3/STAT5B variants with neoplastic cells identified by flow cytometry and genetic features in STAT3/STAT5B-mutant LGLLs and MNs. (A) VAFs of STAT3/STAT5B variants correlated with their corresponding percentage of atypical T or NK cells detected by flow cytometry (R2 = 0.92) in LGLLs whereas there seemed to be no such a correlation in those in (B) MNs (R2 = 0.12). (C) In patients with LGLLs, 35% had concomitant somatic pathogenic and likely pathogenic variants, whereas in patients with MNs, 92% (P < .0001) had at least 1 concomitant variant beyond STAT3/STAT5B variants. (D) The average number of somatic variants was significantly lower in patients with LGLLs (median, 1.7 per patient; range, 1-4 per patient) than in patients with MNs (median, 4.2 per patient; range, 1-8 per patient; ∗∗∗∗P < .0001). (E) The VAFs of the leading concomitant (beyond STAT3/STAT5B) variant in LGLLs (mean, 11.9, range 1 to 32) seemed to be significantly smaller than that in MNs (mean, 37.5, range 2 to 98; ∗∗∗ P = .0003). (F) In LGLLs, there seemed to be a significant correlation (R2= 0.90) between the VAFs of STAT3/STAT5B variants to the VAF of the leading non- STAT3/STAT5B variants, suggesting that STAT3/STAT5B variants are the leading variants in all patients with LGLLs (100% in panel H, blue). (G) This correlation did not exist in MNs (R2 = 0.20), and indeed the data suggested that 48% of all STAT3/STAT5B variants (above the dashed line in panel G) were a subclone (panel H; red; P < .0001).
Correlation of STAT3/STAT5B variants with neoplastic cells identified by flow cytometry and genetic features in STAT3/STAT5B-mutant LGLLs and MNs. (A) VAFs of STAT3/STAT5B variants correlated with their corresponding percentage of atypical T or NK cells detected by flow cytometry (R2 = 0.92) in LGLLs whereas there seemed to be no such a correlation in those in (B) MNs (R2 = 0.12). (C) In patients with LGLLs, 35% had concomitant somatic pathogenic and likely pathogenic variants, whereas in patients with MNs, 92% (P < .0001) had at least 1 concomitant variant beyond STAT3/STAT5B variants. (D) The average number of somatic variants was significantly lower in patients with LGLLs (median, 1.7 per patient; range, 1-4 per patient) than in patients with MNs (median, 4.2 per patient; range, 1-8 per patient; ∗∗∗∗P < .0001). (E) The VAFs of the leading concomitant (beyond STAT3/STAT5B) variant in LGLLs (mean, 11.9, range 1 to 32) seemed to be significantly smaller than that in MNs (mean, 37.5, range 2 to 98; ∗∗∗ P = .0003). (F) In LGLLs, there seemed to be a significant correlation (R2= 0.90) between the VAFs of STAT3/STAT5B variants to the VAF of the leading non- STAT3/STAT5B variants, suggesting that STAT3/STAT5B variants are the leading variants in all patients with LGLLs (100% in panel H, blue). (G) This correlation did not exist in MNs (R2 = 0.20), and indeed the data suggested that 48% of all STAT3/STAT5B variants (above the dashed line in panel G) were a subclone (panel H; red; P < .0001).
Cytogenetics showed predominantly normal or low-risk karyotypes among LGLLs (64%) and a greater proportion of complex karyotypes among MNs (36% in LGLLs vs 64% in MNs with complex karyotypes; P = .01; Figure 1A). T-cell clonality was uniformly positive among patients with T-LGLLs and other T-LPDs and was uniformly negative in patients with NK-LGLLs and MNs in whom it was performed, with 2 exceptions of patients with composite T-LGLL and MDS (patients 66 and 67 in Figure 1A and supplemental Table 3). Rare patients with cytopenia who had STAT3 variants (patients 69-76 in Figure 1A and supplemental Table 3) had mutational profiles more like those of patients with LGLLs, among whom T-cell clonality was negative in 2 patients and unknown in the remaining 5. Given that mutations involving epigenetic regulation are common in both clonal cytopenia of undetermined significance and in LGLLs,22,23 diligent evaluation of T-cell clonality accompanied by flow cytometric studies could be helpful in avoiding missed diagnoses of LGLLs in such patients.
This study was limited by the lack of a subgroup analysis of patients with LGLLs vs patients with STAT3/STAT5B-mutant MDS with known, persistent clonal LGL expansions8,15 because only 3 such patients were identified (patients 89, 90, and 107; supplemental Table 3) whereas attempts to assess T-cell clonality provided no further support. Such a study would require prospective assessments of T-cell clonality and possibly cell sorting in all STAT3/STAT5B-mutated MNs with clonal LGL expansions.10,23,24 Although a myeloid NGS panel is often sufficient when evaluating cytopenias, use of an expanded panel that includes genes whose mutations are common in LNs22 would provide more information for classification, especially in patients with composite LGLLs and MNs.10,15
In conclusion, our study is the first to characterize the molecular features of STAT3/STAT5B-mutant LGLLs and MNs by using a myeloid malignancy–targeted NGS panel. This study supports NGS as a necessary test for facilitating an accurate diagnosis of LGLLs vs MNs in patients with diagnostic challenges. Summaries of genetic features that distinguish LGLLs from MNs include more frequent STAT3 mutations, a lower median VAF of STAT3/STAT5B variants, the absence or reduced diversity of concomitant somatic variants, and STAT3/STAT5B variants being the founder and leading clonal driver variant.
Contribution: P.L. designed the study; P.L. and M.K. performed data and statistical analysis; M.K. drafted the manuscript; P.L., M.M., M.R.A., W.X., P.W.R., G.Y., and W.C. collected patients’ clinical, flow cytometric, cytogenetic, and molecular data; P.L., M.M., M.R.A., W.X., P.W.R., R.D.P., G.Y., and W.C. interpreted and classified all variants by NGS; and all authors reviewed and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peng Li, Associated Regional and University Pathologists (ARUP) Laboratories, 500 Chipeta Way, Salt Lake City, UT 84108; e-mail: .
References
Author notes
Please contact Peng Li at peng.li@aruplab.com or peng.li@hsc.utah.edu for data sharing.
The full-text version of this letter contains a data supplement.

![Comprehensive genetic profiles, hotspots, and VAF distribution of STAT3 and STAT5B variants in LNs and MNs. (A) LNs are displayed on the left (light blue, top row) and MNs on the right (light orange, top row). The World Health Organization (WHO) entities in each group are listed in the second row. The concomitant variants were grouped into the following 6 categories on the basis of their gene function: Epigenetic: epigenetic regulators, genes involving DNA methylation or histone acetylation and deacetylation (light green); SFs: RNA splicing factors (purple); TFs: transcription factors (orange); Signaling: molecules in tyrosine kinase pathway or RAS/MAPK pathways (pink); C: cohesins (light purple); and Others: genes with functions beyond the above categories (various colors). Each column represents 1 patient, each bar represents 1 variant, and split bars indicated 2 or more variants in the same gene. (B) LNs showed predominantly STAT3 variants compared with (C) STAT5B variants (P = .0003), and variants seemed to be concentrated in the SH2 domain of both genes. Among STAT3 variants, in LNs, the D661 variants (including both D661V and D661Y) were most prevalent followed by Y640F. In contrast, in MNs, D661Y, not D661V, was the most common hotspot mutation followed by S614R and Y640F. (C) The STAT5BN642H variant was particularly represented among MNs and only rarely detected in LNs. (D) The distribution of VAFs among LGLLs peaked at a median of 8.8% (solid line; range, 1.4%-48.6%). (E) The VAF distribution in MNs seemed wider with a median VAF of 12.0% (solid line; range, 1.1%-65.2%; [∗] P = .01 by unpaired Student t test) and a significant second population concentrated around 48% to 50% (P = .006 by χ2 test for trend). The dashed lines represent the 25th and 75th percentiles. The relative density of VAF distribution is displayed as previously described.21 AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; MPN, myeloproliferative neoplasm; Neg, negative; NI, no information; NL, normal; NOS, not otherwise specified; SH2, Src homology 2 domain; TAD, topologically associating domain.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/1/10.1182_bloodadvances.2022008192/6/m_blooda_adv-2022-008192-gr1a.jpeg?Expires=1769193416&Signature=qtJAFzsIWk-OINjPdeV74nJ-BB2iiisLHGhDczs5PgAAgf0BFig5HlDEno9fbCOnUdTGkvujQTbU2Po9n13VW2V0ifGCwiGk7E5pX2ICce-w6mAiYCLKNqZtAGAUAycpBte9R78UwJQts5EH9~jP0gmkhXC8ei~EEg0HaXX0ZwoMfk4HzAZmA-jv4MchBtjJb95-do2YPAN39-nHLopkdXhSzebt~rQ5-e7H-4Ff~RPZ82wH-YkG32PNntxUHzBm0RIobSdEIncuEgLrCTIdS1cx1R~Xl5zd8KDf0iLa7f6YDjY3zs13F4GrWMCIbE8LXLRXyeP2cKW3qZTXQG-fEw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Comprehensive genetic profiles, hotspots, and VAF distribution of STAT3 and STAT5B variants in LNs and MNs. (A) LNs are displayed on the left (light blue, top row) and MNs on the right (light orange, top row). The World Health Organization (WHO) entities in each group are listed in the second row. The concomitant variants were grouped into the following 6 categories on the basis of their gene function: Epigenetic: epigenetic regulators, genes involving DNA methylation or histone acetylation and deacetylation (light green); SFs: RNA splicing factors (purple); TFs: transcription factors (orange); Signaling: molecules in tyrosine kinase pathway or RAS/MAPK pathways (pink); C: cohesins (light purple); and Others: genes with functions beyond the above categories (various colors). Each column represents 1 patient, each bar represents 1 variant, and split bars indicated 2 or more variants in the same gene. (B) LNs showed predominantly STAT3 variants compared with (C) STAT5B variants (P = .0003), and variants seemed to be concentrated in the SH2 domain of both genes. Among STAT3 variants, in LNs, the D661 variants (including both D661V and D661Y) were most prevalent followed by Y640F. In contrast, in MNs, D661Y, not D661V, was the most common hotspot mutation followed by S614R and Y640F. (C) The STAT5BN642H variant was particularly represented among MNs and only rarely detected in LNs. (D) The distribution of VAFs among LGLLs peaked at a median of 8.8% (solid line; range, 1.4%-48.6%). (E) The VAF distribution in MNs seemed wider with a median VAF of 12.0% (solid line; range, 1.1%-65.2%; [∗] P = .01 by unpaired Student t test) and a significant second population concentrated around 48% to 50% (P = .006 by χ2 test for trend). The dashed lines represent the 25th and 75th percentiles. The relative density of VAF distribution is displayed as previously described.21 AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; MPN, myeloproliferative neoplasm; Neg, negative; NI, no information; NL, normal; NOS, not otherwise specified; SH2, Src homology 2 domain; TAD, topologically associating domain.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/1/10.1182_bloodadvances.2022008192/6/m_blooda_adv-2022-008192-gr1b.jpeg?Expires=1769193416&Signature=xAaUsN4rt-W-kvjF9xllqOFzXRnulvEmXLrmZ8hKHfXqpO1VLKKc10wsS1Rwp-9Yf30tjsnUPrcIRdMjAs~3T2RabVLOFkje5iX~4bQ6GoJUwxdzDi2LEO1N8LCPlXBrzPaT-1uMgFgKH3Brca0BPphbYEK1W2Paz98n8ug0WOrzOlxvM907gpSKcpuNnliM3UPSurytNl3fxl2pdxatVDnJYlQZxdm56mFXFTjYEtrg74a1bwLfyOR7FsCwqqt1WWZhNpKZwDG7x4nOWDjRzHEpwEA5qiouNITYQVea7TqC7QofxtL2fagZ1f4QP0u5a9aXZEAcJK1EYttgsiTagg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)