Key Points
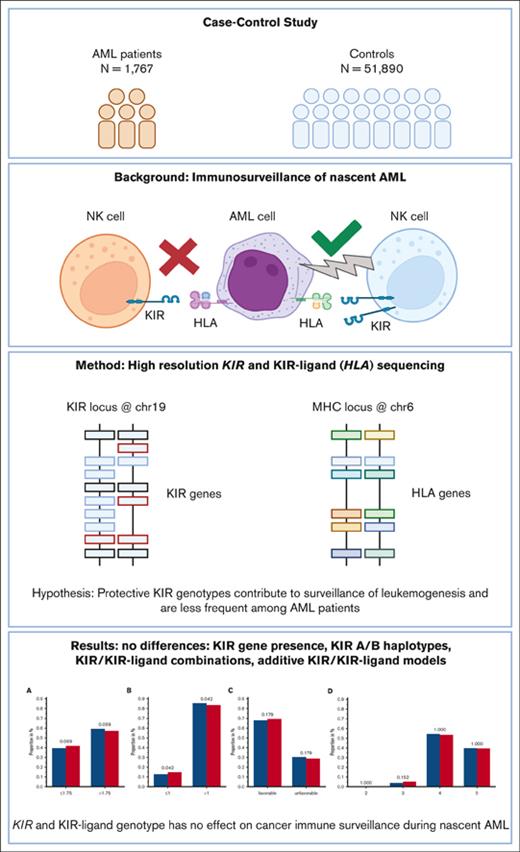
This large case-control study reveals no differences in the KIR and KIR-ligand genotype frequencies among patients with and without AML.
Results suggest steady-state KIR-mediated NK cell immunity does not define risk of AML, and NK immunity is disarmed as leukemic clones grow.
Abstract
Immunogenetic association studies may give rise to new hypotheses on the immune surveillance of cancer. We hypothesized that certain combinations of killer immunoglobulin-like receptor (KIR) and HLA genotypes may enhance natural killer (NK) cell immunity against nascent acute myeloid leukemia (AML) and, thereby, lead to a skewed genotype distribution among patients. For this purpose, we analyzed KIR and HLA genotypes of 1767 German patients with AML and compared the results with that of the data of 51 890 German volunteers who had registered with German bone marrow donor file (DKMS). Patient samples were retrieved from the Collaborative Biobank and the biorepository of the Study Alliance Leukemia. All samples were genotyped with high-resolution amplicon-based next-generation sequencing. Because of the large number of controls, this study was very sensitive to detect the impact of KIR genotype. Knowledge on KIRs and their cognate HLA ligands allowed for testing of several hypotheses of NK cell–mediated endogenous leukemia surveillance. We did not find significant differences between the 2 cohorts in regard to the presence or absence of single KIR genes. When grouped based on telomeric or centromeric gene content, the major haplotypes A/A, A/B, and B/B were equally distributed among patients and control subjects. Using information on KIRs and their HLA ligands, we further tested receptor-ligand models and summation models without revealing markedly significant differences between patients and controls, albeit we observed a trend pointing at a minor protective effect of a low number of inhibitory KIR/KIR-ligand pairs. The results suggest that the KIR/KIR-ligand genotype has no effect on the susceptibility for the development of de novo AML.
Introduction
Natural killer (NK) cells have the potential to kill leukemic cells and may therefore be critical for the endogenous surveillance of leukemogenesis by the immune system. Antileukemic activity of NK cells might be triggered by the loss of HLA, alteration in HLA expression, upregulation of stress molecules (such as HLA-G), or stress-induced (open) conformations of HLA molecules (eg, HLA-F) resulting from enhanced proliferative, oxidative, or immune attack–mediated stress.1,2 Upon the binding of NK cell receptors to their cognate ligands on target cells, intracellular signals modulate the NK cell’s cytotoxic function. In this context killer immunoglobulin-like receptor (KIR) proteins play a pivotal role. KIR molecules comprise a family of activating and inhibitory receptors. Known ligands for most of the inhibitory KIRs (iKIRs) are specific sets of HLA class I molecules. Some activating KIRs also bind to selected HLA class I molecules but with lower affinity, in a peptide-dependent manner and stress-induced conformation or expression.3-5KIR genes are encoded on chromosome 19. In an individual, KIR genes might be present or absent, can differ in gene copy number, and display a distinct allelic polymorphism.6 The A and B haplotypes are the 2 major KIR haplotypes, with A containing a fixed number of predominantly inhibitory receptors and B having both inhibiting and activating KIR receptors. Furthermore, the KIR genes display an extensive allelic polymorphism, with between 33 alleles (for KIR2DS1) and 228 alleles (for KIR3DL3) identified so far. For 9 KIR genes, null alleles have been described, not being expressed at the cell surface.7
We aim to test the hypothesis that KIR genotypes vary in their potential of endogenous immune surveillance of the development of acute myeloid leukemia (AML). In recent studies, the presence or absence of certain KIR genes, KIR haplotype motifs, or KIR/KIR-ligand combinations have been reported to be associated with the susceptibility to breast cancer,8,9 colorectal cancer,10,11 or other solid tumors.12,13 Associations of KIR genotypes with the risk of developing AML were investigated in small case-control studies,14-18 and the results were not consistent.
Models have been proposed to explain the contribution of KIRs to the susceptibility to virus infections,19 tumors,10 autoimmunity,20 and relapse after hematopoietic stem cell transplantation.21 Each of these challenges to the immune system has unique features. Yet, testing these models for susceptibility to de novo AML could shed light on a potential role for KIR-mediated endogenous NK cell immune surveillance. These models have been developed to classify KIRs together with their cognate ligands into favorable or unfavorable combinations theoretically conferring more or less potent NK cell effector activity.21-23 As an example, donors bearing either A or B haplotypes were found to be associated with susceptibility to viral infections24,25 or cancer26,27; and donors with B haplotypes were found to confer an improved outcome in unrelated stem cell transplantations.21,28 Supposedly, favorable KIR genotypes may also be protective during leukemogenesis; in the lifetime of every human being, hematopoietic stem cells acquire somatic mutations,29 and some of these cells may eventually transform into malignant cells. The immune system of the affected individual has the potential to detect and to eliminate these cells. T cells might fight leukemic cells when the latter are displaying tumor-associated or tumor-specific peptide antigens via HLA molecules. Malignant myeloid cells, in turn, may circumvent T-cell immune surveillance via HLA expression reduction30 or HLA loss of heterozygosity already at diagnosis.31 On the other hand, this mechanism of immune evasion might be recognized by NK cells, for examples, via iKIRs. Many activating KIRs may also contribute to superior leukemic cell killing. The effective control of the immune system limits the lifetime risk of AML to <1%. One could expect that favorable KIR genotypes contribute to endogenous immune surveillance of nascent AML and are, therefore, less frequent among patients with AML compared with healthy individuals, whereas KIR genotypes permissive to leukemogenesis are more frequent among patients with AML than among healthy individuals. To test this hypothesis, we performed a case-control study comparing KIR genotypes of 1689 patients with AML with those of 51 890 healthy individuals.
Methods
Samples from patients and donors
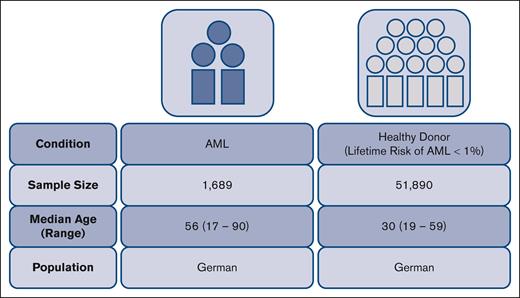
We conducted a case-control study on the impact of KIR genotype information on patients with AML who contributed a sample to the Collaborative Biobank or Study Alliance Leukemia biobank. All patients, whose samples were analyzed in this study, had provided written informed consent when they contributed a sample to the respective biobank. Additional eligibility criteria were a diagnosis of AML and an age of ≥18 years. Patients’ characteristics are compiled in Table 1. Immunogenetic data for the reference group were taken from recently typed individuals who had registered as potential stem cell donors with German bone marrow donor file (DKMS). Both, donors and patients are German cohorts (Figure 1). The study was approved by the ethical committee at the Technical University of Dresden (EK 249072018).
Patients’ characteristics
| . | Patients with AML . | Healthy control group . |
|---|---|---|
| Demographic data | ||
| Median age (range), y∗ | 56 (17-90) | 30 (19-59) |
| Sex, %∗ | ||
| Female | 49.5 | 65.9 |
| Male | 50.5 | 34.1 |
| Population | German | German |
| Biobank, n | ||
| Collaborative Biobank | 505 | |
| Study Alliance Leukemia | 1265 | |
| Disease classification, %∗ | ||
| AML status | ||
| De novo AML | 88.9 | |
| Secondary AML | 10.4 | |
| Tertiary AML | 0.7 | |
| Disease status at the time of sample collection, % | ||
| Initial AML diagnosis | 100 | |
| European LeukemiaNet risk category, % | ||
| Favorable | 37.7 | |
| Intermediate | 57.4 | |
| Adverse | 4.7 | |
| Not applicable | 0.2 | |
| Eastern Cooperative Oncology Group, % | ||
| 0 | 20.2 | |
| 1 | 51.1 | |
| 2 | 16.8 | |
| 3 | 3.9 | |
| 4 | 1.4 | |
| Not applicable | 6.6 | |
| . | Patients with AML . | Healthy control group . |
|---|---|---|
| Demographic data | ||
| Median age (range), y∗ | 56 (17-90) | 30 (19-59) |
| Sex, %∗ | ||
| Female | 49.5 | 65.9 |
| Male | 50.5 | 34.1 |
| Population | German | German |
| Biobank, n | ||
| Collaborative Biobank | 505 | |
| Study Alliance Leukemia | 1265 | |
| Disease classification, %∗ | ||
| AML status | ||
| De novo AML | 88.9 | |
| Secondary AML | 10.4 | |
| Tertiary AML | 0.7 | |
| Disease status at the time of sample collection, % | ||
| Initial AML diagnosis | 100 | |
| European LeukemiaNet risk category, % | ||
| Favorable | 37.7 | |
| Intermediate | 57.4 | |
| Adverse | 4.7 | |
| Not applicable | 0.2 | |
| Eastern Cooperative Oncology Group, % | ||
| 0 | 20.2 | |
| 1 | 51.1 | |
| 2 | 16.8 | |
| 3 | 3.9 | |
| 4 | 1.4 | |
| Not applicable | 6.6 | |
Patient data only available for Study Alliance Leukemia biobank samples.
HLA and KIR genotyping
At the time of registration to DKMS, volunteers provided buccal swabs or blood for DNA extraction and genotyping. The standard genotyping of a panel of genes relevant for stem cell donor selection, including information on HLA-A, -B, -C, or -DRB1 was predominantly performed at the DKMS Life Science Lab, applying a high-resolution amplicon-based approach, using Illumina devices.32KIR genotyping was performed using a high-resolution short-amplicon–based next-generation sequencing workflow. KIR typing at the allele level was based on sequencing of exons 3, 4, 5, 7, 8, and 9 and subsequent bioinformatic analysis, as described previously.33 DNA of patients and donors were typed with the same workflow.
Data preparation
Typing of 78 of the 1767 patient samples failed because of insufficient DNA concentration or quality for the workflow. The final analysis set thus contained information on 1689 patients. HLA-C alleles were grouped in C1 and C2 ligands, and B alleles were grouped into Bw4-80I/Bw-80T/Bw6 epitopes based on information retrieved. Expression levels of KIR3DL1 alleles were classified per the criteria by Boudreau et al, into high, low, or null.34 Further information on KIR3DL1 and KIR2DS1 and their cognate HLA class I ligands was used to group patients and donors in accordance with the criteria presented in the studies by Venstrom et al23 and Boudreau et al.22 For calculating KIR/KIR-ligand scores, models described by Boelen et al,19 Rafei et al,35 and Krieger et al36 were applied. KIR absence/presence data were available for 99.95% of the study population. Because of the remaining ambiguity testing for KIR3DL1 interaction, models were restricted to 1469 cases. The numbers of cases evaluated for each test are reported in parentheses in Tables 2 and 3.
KIR gene presence, KIR haplotype motif-based models, and models based on KIR/KIR-ligand interactions
| Classifier . | No. of patients with pos AML (No. of total patients with AML) . | Portion of patients with pos AML . | No. of pos control subjects (No. of total control subjects) . | Portion of pos control group . | P . | Adjusted P . |
|---|---|---|---|---|---|---|
| KIRgene presence | ||||||
| KIR2DL1 | 1 640 (1 689) | 0.971 | 50 195 (51 885) | 0.967 | .458 | 1.000 |
| KIR2DL2 | 896 (1 689) | 0.530 | 26 802 (51 888) | 0.517 | .269 | 1.000 |
| KIR2DL3 | 1 511 (1 689) | 0.894 | 46 956 (51 888) | 0.905 | .167 | 1.000 |
| KIR2DL5 | 832 (1 689) | 0.493 | 25 353 (51 890) | 0.489 | .765 | 1.000 |
| KIR2DP1 | 1 643 (1 689) | 0.973 | 50 344 (51 887) | 0.970 | .600 | 1.000 |
| KIR2DS1 | 635 (1 689) | 0.376 | 19 927 (51 890) | 0.384 | .519 | 1.000 |
| KIR2DS2 | 898 (1 689) | 0.532 | 27 053 (51 888) | 0.521 | .418 | 1.000 |
| KIR2DS3 | 495 (1 689) | 0.293 | 14 928 (51 890) | 0.288 | .650 | 1.000 |
| KIR2DS4 | 1 617 (1 677) | 0.964 | 49 347 (51 890) | 0.951 | .015 | .180 |
| KIR2DS5 | 483 (1 689) | 0.286 | 15 013 (51 890) | 0.289 | .785 | 1.000 |
| KIR3DL1 | 1 617 (1 689) | 0.957 | 49 354 (51 887) | 0.951 | .268 | 1.000 |
| KIR3DS1 | 630 (1 689) | 0.373 | 19 535 (51 888) | 0.376 | .786 | 1.000 |
| Cen A/B haplotypes, overall χ2testP = .332 | ||||||
| Cen A/A | 789 (1 689) | 0.467 | 24 780 (51 888) | 0.478 | .412 | 1.000 |
| Cen A/B | 722 (1 689) | 0.427 | 22 176 (51 888) | 0.427 | 1.000 | 1.000 |
| Cen B/B | 178 (1 689) | 0.105 | 4 932 (51 888) | 0.095 | .167 | .502 |
| Tel A/B haplotypes, overall χ2testP = .487 | ||||||
| Tel A/A | 1 019 (1 689) | 0.603 | 31 071 (51 889) | 0.599 | .728 | 1.000 |
| Tel A/B | 598 (1 689) | 0.354 | 18 275 (51 889) | 0.352 | .895 | 1.000 |
| Tel B/B | 72 (1 689) | 0.043 | 2 543 (51 889) | 0.049 | .254 | .763 |
| B content score, overall χ2testP = .993 | ||||||
| B content 0 | 535 (1 689) | 0.317 | 16 659 (51 890) | 0.321 | .730 | 1.000 |
| B content 1 | 638 (1 689) | 0.378 | 19 528 (51 890) | 0.376 | .927 | 1.000 |
| B content 2 | 382 (1 689) | 0.226 | 11 715 (51 890) | 0.226 | .992 | 1.000 |
| B content 3 | 118 (1 689) | 0.070 | 3 509 (51 890) | 0.068 | .775 | 1.000 |
| B content 4 | 16 (1 689) | 0.009 | 479 (51 890) | 0.009 | 1.000 | 1.000 |
| KIR3DL1/HLA-B subtype combinations, overall χ2testP = .402 | ||||||
| Strong inhibiting KIR3DL1 | 382 (1 469) | 0.260 | 11 928 (46 375) | 0.257 | .830 | 1.000 |
| Weak inhibiting KIR3DL1 | 406 (1 469) | 0.276 | 12 133 (46 375) | 0.262 | .217 | .867 |
| Educated, uninhibited KIR3DL1 | 118 (1 469) | 0.080 | 4 172 (46 375) | 0.090 | .220 | .880 |
| Missing ligand KIR3DL1 | 563 (1 469) | 0.383 | 18 142 (46 375) | 0.391 | .557 | 1.000 |
| KIR2DS1/C1C2 epitope combinations, overall χ2testP = .186 | ||||||
| KIR2DS1 neg | 1 054 (1 666) | 0.633 | 31 963 (50 619) | 0.631 | .940 | 1.000 |
| KIR2DS1 pos, C1+ | 521 (1 666) | 0.313 | 16 346 (50 619) | 0.323 | .396 | 1.000 |
| KIR2DS1 pos, C2/C2 | 91 (1 666) | 0.055 | 2 310 (50 619) | 0.046 | .096 | .288 |
| Classifier . | No. of patients with pos AML (No. of total patients with AML) . | Portion of patients with pos AML . | No. of pos control subjects (No. of total control subjects) . | Portion of pos control group . | P . | Adjusted P . |
|---|---|---|---|---|---|---|
| KIRgene presence | ||||||
| KIR2DL1 | 1 640 (1 689) | 0.971 | 50 195 (51 885) | 0.967 | .458 | 1.000 |
| KIR2DL2 | 896 (1 689) | 0.530 | 26 802 (51 888) | 0.517 | .269 | 1.000 |
| KIR2DL3 | 1 511 (1 689) | 0.894 | 46 956 (51 888) | 0.905 | .167 | 1.000 |
| KIR2DL5 | 832 (1 689) | 0.493 | 25 353 (51 890) | 0.489 | .765 | 1.000 |
| KIR2DP1 | 1 643 (1 689) | 0.973 | 50 344 (51 887) | 0.970 | .600 | 1.000 |
| KIR2DS1 | 635 (1 689) | 0.376 | 19 927 (51 890) | 0.384 | .519 | 1.000 |
| KIR2DS2 | 898 (1 689) | 0.532 | 27 053 (51 888) | 0.521 | .418 | 1.000 |
| KIR2DS3 | 495 (1 689) | 0.293 | 14 928 (51 890) | 0.288 | .650 | 1.000 |
| KIR2DS4 | 1 617 (1 677) | 0.964 | 49 347 (51 890) | 0.951 | .015 | .180 |
| KIR2DS5 | 483 (1 689) | 0.286 | 15 013 (51 890) | 0.289 | .785 | 1.000 |
| KIR3DL1 | 1 617 (1 689) | 0.957 | 49 354 (51 887) | 0.951 | .268 | 1.000 |
| KIR3DS1 | 630 (1 689) | 0.373 | 19 535 (51 888) | 0.376 | .786 | 1.000 |
| Cen A/B haplotypes, overall χ2testP = .332 | ||||||
| Cen A/A | 789 (1 689) | 0.467 | 24 780 (51 888) | 0.478 | .412 | 1.000 |
| Cen A/B | 722 (1 689) | 0.427 | 22 176 (51 888) | 0.427 | 1.000 | 1.000 |
| Cen B/B | 178 (1 689) | 0.105 | 4 932 (51 888) | 0.095 | .167 | .502 |
| Tel A/B haplotypes, overall χ2testP = .487 | ||||||
| Tel A/A | 1 019 (1 689) | 0.603 | 31 071 (51 889) | 0.599 | .728 | 1.000 |
| Tel A/B | 598 (1 689) | 0.354 | 18 275 (51 889) | 0.352 | .895 | 1.000 |
| Tel B/B | 72 (1 689) | 0.043 | 2 543 (51 889) | 0.049 | .254 | .763 |
| B content score, overall χ2testP = .993 | ||||||
| B content 0 | 535 (1 689) | 0.317 | 16 659 (51 890) | 0.321 | .730 | 1.000 |
| B content 1 | 638 (1 689) | 0.378 | 19 528 (51 890) | 0.376 | .927 | 1.000 |
| B content 2 | 382 (1 689) | 0.226 | 11 715 (51 890) | 0.226 | .992 | 1.000 |
| B content 3 | 118 (1 689) | 0.070 | 3 509 (51 890) | 0.068 | .775 | 1.000 |
| B content 4 | 16 (1 689) | 0.009 | 479 (51 890) | 0.009 | 1.000 | 1.000 |
| KIR3DL1/HLA-B subtype combinations, overall χ2testP = .402 | ||||||
| Strong inhibiting KIR3DL1 | 382 (1 469) | 0.260 | 11 928 (46 375) | 0.257 | .830 | 1.000 |
| Weak inhibiting KIR3DL1 | 406 (1 469) | 0.276 | 12 133 (46 375) | 0.262 | .217 | .867 |
| Educated, uninhibited KIR3DL1 | 118 (1 469) | 0.080 | 4 172 (46 375) | 0.090 | .220 | .880 |
| Missing ligand KIR3DL1 | 563 (1 469) | 0.383 | 18 142 (46 375) | 0.391 | .557 | 1.000 |
| KIR2DS1/C1C2 epitope combinations, overall χ2testP = .186 | ||||||
| KIR2DS1 neg | 1 054 (1 666) | 0.633 | 31 963 (50 619) | 0.631 | .940 | 1.000 |
| KIR2DS1 pos, C1+ | 521 (1 666) | 0.313 | 16 346 (50 619) | 0.323 | .396 | 1.000 |
| KIR2DS1 pos, C2/C2 | 91 (1 666) | 0.055 | 2 310 (50 619) | 0.046 | .096 | .288 |
The framework genes KIR3DL3, 3DP1, 2DL4, and 3DL2 are present in all genomes of the patients with AML and in >99.94% of the healthy individuals, and therefore, not compiled in this table. KIR2DS4 is counted ‘present’ when either the full-length or the truncated variant of the allele or both are present at gene level. Numbers of cases evaluated for each test vary because of remaining ambiguities and are reported in parentheses.
Cen, centromeric; neg, negative; pos, positive; Tel, telomeric.
Additive KIR/KIR-ligand models
| Classifier . | No. of respective patients with AML (No. of total patients with AML) . | Portion of respective patients with AML . | No. of respective control subjects (No. of total control subjects) . | Portion of respective control subjects . | P . | Adjusted P . |
|---|---|---|---|---|---|---|
| Inhibitory KIR/KIR-ligand count considering KIR3DL1/HLA-Bw4 (Boelen et al), overall χ2 test P = .042 | ||||||
| Functional iKIR count (cutoff ≤ 1) | 229 (1 676) | 0.137 | 7 743 (49 898) | 0.155 | .042 | |
| Functional iKIR count (cutoff > 1) | 1 447 (1 676) | 0.863 | 42 155 (49 898) | 0.845 | ||
| Inhibitory KIR/KIR-ligand score considering KIR3DL1/HLA-Bw4 (Boelen et al), overall χ2 test P = .079 | ||||||
| Inhibitory score (cutoff ≤ 1.75) | 674 (1 676) | 0.402 | 21 157 (49 898) | 0.424 | .079 | |
| Inhibitory score (cutoff > 1.75) | 1 002 (1 676) | 0.598 | 28 741 (49 898) | 0.576 | ||
| Inhibitory and activating KIR/KIR-ligand matches∗(Rafei et al), overall χ2 test P = .164 | ||||||
| Inhibitory and activating favorable | 1 087 (1 581) | 0.688 | 29 695 (42 171) | 0.704 | .164 | |
| Inhibitory and activating unfavorable† | 494 (1 581) | 0.312 | 12 476 (42 171) | 0.296 | ||
| Nonweighted additive inhibitory/missing-ligand KIR score (Krieger et al), overall χ2 test P = .206 | ||||||
| 2 | 2 (1 584) | 0.001 | 61 (42 177) | 0.001 | 1.000 | 1.000 |
| 3 | 73 (1 584) | 0.046 | 2 483 (42 177) | 0.059 | .039 | .158 |
| 4 | 870 (1 584) | 0.550 | 22 780 (42 177) | 0.540 | .440 | 1.000 |
| 5 | 636 (1 584) | 0.402 | 16 853 (42 177) | 0.400 | .850 | 1.000 |
| Classifier . | No. of respective patients with AML (No. of total patients with AML) . | Portion of respective patients with AML . | No. of respective control subjects (No. of total control subjects) . | Portion of respective control subjects . | P . | Adjusted P . |
|---|---|---|---|---|---|---|
| Inhibitory KIR/KIR-ligand count considering KIR3DL1/HLA-Bw4 (Boelen et al), overall χ2 test P = .042 | ||||||
| Functional iKIR count (cutoff ≤ 1) | 229 (1 676) | 0.137 | 7 743 (49 898) | 0.155 | .042 | |
| Functional iKIR count (cutoff > 1) | 1 447 (1 676) | 0.863 | 42 155 (49 898) | 0.845 | ||
| Inhibitory KIR/KIR-ligand score considering KIR3DL1/HLA-Bw4 (Boelen et al), overall χ2 test P = .079 | ||||||
| Inhibitory score (cutoff ≤ 1.75) | 674 (1 676) | 0.402 | 21 157 (49 898) | 0.424 | .079 | |
| Inhibitory score (cutoff > 1.75) | 1 002 (1 676) | 0.598 | 28 741 (49 898) | 0.576 | ||
| Inhibitory and activating KIR/KIR-ligand matches∗(Rafei et al), overall χ2 test P = .164 | ||||||
| Inhibitory and activating favorable | 1 087 (1 581) | 0.688 | 29 695 (42 171) | 0.704 | .164 | |
| Inhibitory and activating unfavorable† | 494 (1 581) | 0.312 | 12 476 (42 171) | 0.296 | ||
| Nonweighted additive inhibitory/missing-ligand KIR score (Krieger et al), overall χ2 test P = .206 | ||||||
| 2 | 2 (1 584) | 0.001 | 61 (42 177) | 0.001 | 1.000 | 1.000 |
| 3 | 73 (1 584) | 0.046 | 2 483 (42 177) | 0.059 | .039 | .158 |
| 4 | 870 (1 584) | 0.550 | 22 780 (42 177) | 0.540 | .440 | 1.000 |
| 5 | 636 (1 584) | 0.402 | 16 853 (42 177) | 0.400 | .850 | 1.000 |
The truncated version of KIR2DS4 is not considered as an activating KIR.
≥3 inhibitory KIR/KIR-ligand matches and no activating KIR/KIR-ligand match. Numbers of cases evaluated for each test vary because of remaining ambiguities and are reported in parentheses.
Statistical analysis
For each hypothesis we assessed, KIR and, if applicable, HLA genotypes were categorized (eg, based on HLA-C ligands C1/C1, C1/C2, or C2/C2). The proportion of each category was calculated for patients with AML and controls. If it was not possible to assign all samples to a category, the denominator of this proportion refers to all assignable cases. Proportions were compared using χ2 tests for the resulting contingency tables. Each category was also tested pairwise between groups (eg, C1/C1 in patients with AML vs C1/C1 in the control group) using a 2-way alternative hypothesis. The P values for pairwise comparisons were adjusted for multiple testing based on the number of categories using Bonferroni correction, that is, in the case of HLA-C ligand combinations, the P values for the individual tests for differences between C1/C1, C1/C2, and C2/C2 were multiplied 3× before assessing significance. For numeric variables, logistic regression models were fitted with the group label (AML or control group) as response and the test variables as sole explanatory variables. Figures were created with GraphPad Prism software and BioRender.com
Power considerations
With the 2 groups, nAML (n=1689) and ncontrol (n=51 890), 2-sided proportion tests of 2 categories at a target power of 80% and a significance level of 0.05, allowed us to detect effect sizes, corresponding to ∼3.5% unit differences at proportions of ∼50%.37 For example, if the proportion of KIR2DL5-present samples in the control group was 0.5, we could detect a proportion >0.535 or <0.465 in the patient group with AML, at 80% power with a 0.05 significance level. In order to monitor the power of χ2 tests for different group sizes, we performed post hoc power calculations for all major comparisons.
Results
Presence or absence of KIR genes
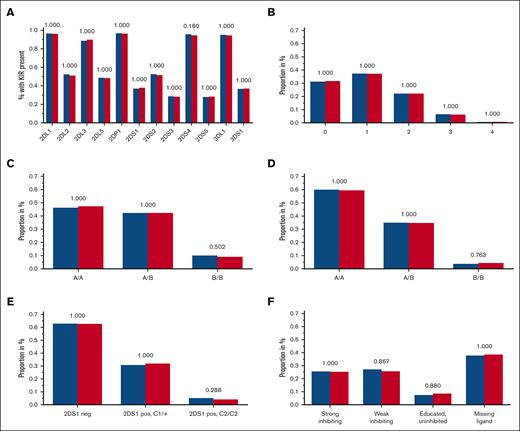
Although the 4 framework KIR genes KIR2DL4, 3DL2, 3DL3, and 3DP1 are present in almost all individuals, each of the other KIR genes is absent in the genome of a considerable portion of healthy subjects and patients with AML. There was no significant difference in the frequency of present KIR2DL1, 2DL2, 2DL3, 2DL5 (A or B), 2DP1, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1, and 3DS1 genes in patients when compared with that in control subjects (Figure 2A). KIR2DS4 was present in 96.4% of patients with AML and 95.1% of the control subjects (χ2 test, unadjusted P = .015: adjusted P = .18). Because published data suggested an association of the 2 KIR2DS4 variant groups, namely full-length and truncated, with leukemia38 and solid tumors,39,40 we analyzed KIR2DS4 in great detail. We did not find significant differences in the distribution of KIR2DS4 full-length or truncated variants (supplemental Table 1; supplemental Figure 1). For the readers’ interest, KIR genotypes for each KIR gene were counted and compiled at the allele level (first-field level) in supplemental Table 2.
Frequencies of tested KIR genotypes. (A-F) Blue columns are displaying frequencies of patients with AML, and red columns are displaying the control group with the respective KIR (or KIR ligand) genotype: KIR gene presence (A), B content score (B), centromeric A/B haplotypes (C), telomeric A/B haplotypes (D), KIR2DS1 and HLA-C1/C2 (E), and KIR3DL1 HLA-B subtype (F) combinations based on the criteria by Boudreau et al22; adjusted P values are depicted above the pairs of columns.
Frequencies of tested KIR genotypes. (A-F) Blue columns are displaying frequencies of patients with AML, and red columns are displaying the control group with the respective KIR (or KIR ligand) genotype: KIR gene presence (A), B content score (B), centromeric A/B haplotypes (C), telomeric A/B haplotypes (D), KIR2DS1 and HLA-C1/C2 (E), and KIR3DL1 HLA-B subtype (F) combinations based on the criteria by Boudreau et al22; adjusted P values are depicted above the pairs of columns.
KIR A/B haplotypes
Putative KIR A/B haplotypes can be defined by different sets of KIR genes; the KIR A haplotype comprises a defined set of iKIR genes and the activating KIR gene KIR2DS4, which is predominantly present as a truncated variant coding for a nonmembrane anchored form in KIR A haplotypes.41 All other KIR haplotypes are assigned to the KIR B haplotypes, which contain more activating KIR genes. We did not find different distributions for haplotype A or B motifs, neither for the centromeric nor for the telomeric region of the KIR locus (Figure 2C-D).
A higher content of centromeric or telomeric B motifs provides NK cells with more activating KIR receptors, which has been associated with increased functional potential and antileukemic activity.21 The number of B motifs can be counted to generate a score ranging from 0 to 4. In our analyses, the distribution of B content scores did not differ between patients and controls (Table 2; Figure 2B).
Presence or absence of KIR-HLA class I ligands
KIR receptors may only modulate the function of NK cells when they bind to their cognate ligands. HLA-C molecules bear either the C1 or the C2 epitope and can be bound by the inhibitory KIR2DL1 (C2), KIR2DL2 (C1), and KIR2DL3 (C1) and, to a lesser extent, also by the activating KIR2DS1 (C2).42 Some HLA-B molecules are carrying the Bw4 epitope, the ligand for KIR3DL1, whereas the Bw6 motif on other HLA-B molecules is not bound. There are 2 subgroups of Bw4 epitopes because of a dimorphism at amino acid position 80 being either isoleucine (I) or threonine (T).
The frequency of C1/C1-positive individuals was higher in the healthy control group (39.4% vs 36.2%; adjusted P = .027), whereas patients with AML exposed C2/C2 more often (14.1% vs 12.4%; adjusted P = .112) (supplemental Table 3; supplemental Figure 2). Individuals homozygous for Bw4-80-I were more frequent in the patient group (3.8% vs 2.5%; adjusted P = .007) (supplemental Table 3; supplemental Figure 3).
KIR/KIR-ligand combinations
Intracellular signaling differs among various KIR3DL1 alleles and depends on the Bw4 dimorphism. Boudreau et al classified patients with AML who had received hematopoietic cell transplantation (HCT) based on their own Bw4 ligands and the donors’ KIR3DL1 alleles in order to predict the risk of relapse.22 They proposed a model with combinations of different alleles of KIR3DL1/KIR3DS1 and their ligands Bw4 80-I or Bw4 80-T, thereby defining the following 4 groups1: educated uninhibiting,2 missing ligand,3 strong inhibiting,4 and weak inhibiting. We did not find any significant differences for these 4 groups when comparing the frequencies in controls and in patients with AML (Table 2; Figure 2F).
The activating KIR2DS1 binds moderately strong to C2, but KIR2DS1+ NK cells become hyporesponsive in C2/C2 individuals.43 Venstrom et al, therefore, grouped patients with AML based on combinations of C1 or C2 ligands and based on their donors’ KIR2DS1 presence or absence to predict the risk of relapse after transplantation.23 We did not find significant different percentages of KIR2DS1–, KIR2DS1+ and C2/C2, and KIR2DS1+ and C1+ individuals among patients and healthy individuals (Table 2; Figure 2E).
Additive KIR/KIR-ligand models
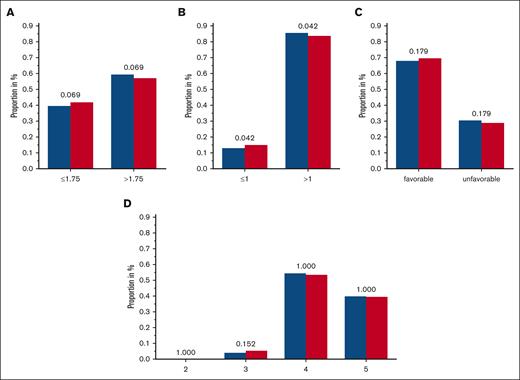
To enlighten the concerted effects that KIRs are supposed to exert, scores were constructed, which integrate the information on KIRs and corresponding KIR ligands. Boelen et al developed 1 such score and demonstrated an impact of different score levels on the course of HIV infections.19 We challenged this and tested it as an ordinal category, as a continuous variable as well as dichotomized with different cutoffs. Both, the comparisons of the functional KIR/KIR-ligand counts and those of the (weighted) KIR/KIR-ligand scores, suggested a marginal protective effect of a low number of inhibitory KIR/KIR-ligand pairs. Specifically, an inhibitory count (resulting from the presence or absence of KIR2DL1, KIR2DL2, KIR2DL3, KIR3DL1, and their cognate ligands) of ≤1 was observed in 13.7% of patients with AML compared with 15.5% of individuals in the control group (unadjusted P = .042) (Table 3 and Figure 3; supplemental Tables 4 and 5; supplemental Figure 4); however, this effect is contrary to those in the findings of the study on viral immunity.19
Frequencies of 3 tested additive KIR/KIR-ligand models. (A-C) Blue columns are displaying frequencies of patients with AML, and red columns are displaying the control group with the respective category of scores/counts of the considered KIR/KIR-ligand-models: inhibitory score based on the findings by Boelen et al,19 including KIR3DL1/HLA-Bw4 and considering KIR2DL2 and KIR2DL3 separately, with a cutoff of 1.75 (A); inhibitory count based on the findings by Boelen et al, including KIR3DL1/HLA-Bw4 and considering KIR2DL2 and KIR2DL3 separately, with a cutoff of 1.0 (B); and favorable and unfavorable score based on the criteria given by Rafei et al35 combining inhibitory and activating scores (C). The truncated version of KIR2DS4 is not considered as an activating KIR; unfavorable is defined by the presence of ≥3 inhibitory KIR/KIR-ligand matches and no activating KIR/KIR-ligand match. (D) Inhibitory/missing-ligand KIR score based on the criteria given by Krieger et al36 accounting for iKIRs and missing KIR ligands; adjusted P values are depicted above the pairs of columns.
Frequencies of 3 tested additive KIR/KIR-ligand models. (A-C) Blue columns are displaying frequencies of patients with AML, and red columns are displaying the control group with the respective category of scores/counts of the considered KIR/KIR-ligand-models: inhibitory score based on the findings by Boelen et al,19 including KIR3DL1/HLA-Bw4 and considering KIR2DL2 and KIR2DL3 separately, with a cutoff of 1.75 (A); inhibitory count based on the findings by Boelen et al, including KIR3DL1/HLA-Bw4 and considering KIR2DL2 and KIR2DL3 separately, with a cutoff of 1.0 (B); and favorable and unfavorable score based on the criteria given by Rafei et al35 combining inhibitory and activating scores (C). The truncated version of KIR2DS4 is not considered as an activating KIR; unfavorable is defined by the presence of ≥3 inhibitory KIR/KIR-ligand matches and no activating KIR/KIR-ligand match. (D) Inhibitory/missing-ligand KIR score based on the criteria given by Krieger et al36 accounting for iKIRs and missing KIR ligands; adjusted P values are depicted above the pairs of columns.
Furthermore, we tested 2 other models developed by Rafei et al35 and Krieger et al,36 both including information on activating and iKIRs and KIR ligands. We could not detect significant differences among patients with AML and control subjects when testing the described scores (Table 3 and Figure 3; supplemental Tables 4 and 5; supplemental Figure 4).
Discussion
The aim of this case-control study is to compare KIR and HLA genotype frequencies among patients with AML and healthy individuals based on the hypothesis that protective KIR and HLA genotypes would be less frequent among patients than among control subjects. We did not find any significant differences in the presence or absence of KIR genes, KIR A/B haplotypes, KIR/KIR-ligand combinations, and additive KIR/KIR-ligand models. Minor differences were detected in the presence or absence of certain KIR-HLA class I ligand pairs, but the presence or absence of KIR genes, which recognize the respective ligands, did not help in understanding this effect.
With 1689 patients, this case-control study, to our knowledge, is 1 of the largest analysis of the impact of KIR genotypes on the development of AML. Because of the high number of 51 890 healthy individuals in the control group, the statistical tests were very sensitive to detect different distributions of genotypes between cases and controls.
One limitation of this study is that the control individuals were younger (median age, 30 years) compared with the patients (median age, 56 years). However, because the lifetime risk of developing AML is only ∼1%, the number of individuals who eventually develop AML later during life would be so small that the genotype distribution of true control subjects would not differ in a meaningful order of magnitude. Moreover, genotype frequencies of the general population usually do not differ substantially between generations, which allows for the comparison of patients and control subjects of different age groups.
We did not find any significant differences when looking for the presence or absence of KIR genes in patients when compared with the control group. Other studies comparing the presence or absence of KIR genes in patients with AML and other hematologic malignancies with that in control groups found differences, but small sample size, heterogeneous groups of patients with leukemia, and/or missing P value correction for multiple testing were limitations of these studies.14-18
Alleles of the KIR2DS4 gene can be grouped based on the full-length or truncated variants, the latter with a 22–base pair deletion in exon 5, coding for a secreted version of the KIR2DS4 protein. Contradictory results have been published, suggesting the presence of full-length KIR2DS4 alleles to be protective or a risk factor in patients with blood cancer. Kamenaric et al published data showing a significantly higher frequency of full-length KIR2DS4 alleles among 111 patients with various hematologic malignancies than among a control group (N = 121).38 In another study, Giebel et al compared the presence and absence of KIR genes in 90 patients with AML, chronic myeloid leukemia, and acute lymphoblastic leukemia, all candidates for unrelated or related HCT, with their HLA-matched donors serving as control subjects.44 The frequency of full-length KIR2DS4∗001 was significantly higher in donors than in all patients with leukemia, but for patients with AML and their respective donors, this difference was not significant. Different results were found referring to the susceptibility to endometrioid cancer observation made by Giebel et al, with KIR2DS4 full-length allele presence being a risk factor.39 We could not find any significantly altered distribution of KIR2DS4 full-length or truncated alleles in patients with AML compared with the control group.
KIR haplotypes comprise a set of KIR genes on 1 chromosome. A simplistic model suggests the assignment of a KIR locus with a defined KIR gene content of mainly iKIRs (KIR2DL3, KIR2DP1, KIR2DL1, KIR3DL1, and all 4 framework genes) and just 1 activating KIR (KIR2DS4) to the A haplotype, whereas all other haplotypes containing more activating KIRs are assigned to the B haplotype. We did not reveal any significant differences in the distribution of KIR A/B haplotypes in patients with AML or in the control group. Others found that KIR haplotype A or B might be associated with leukemia; Deng et al identified a protective effect from developing AML and other hematologic malignancies for KIR haplotype A homozygosity in a Chinese, Han population.45 The authors hypothesized that NK cells were efficiently educated in KIR A/A haplotype–bearing individuals, making these cells particularly capable to detect loss or downregulation of HLA expression or even an altered HLA ligandome displayed by leukemic cells via KIR binding. The KIR A/A haplotype can also be described by a B content score of 0. We found only a minor, nonsignificant difference comparing the frequencies of patients (31.7%) with that of the control group (32.1%), with a B content score of 0. Frequencies of individuals with a KIR A/A haplotype are higher in East Asian populations than the frequencies in European populations or in the German cohort of this study, yet the absolute number of KIR A/A individuals are comparable in both cohorts. However, differences in the frequencies of specific KIR alleles and of HLA class I alleles serving as KIR ligands in various populations might explain the differing results.
On the other hand, in a study including 108 patients with myelodysplastic syndrome, Stringaris et al found that haplotype A is a risk factor for the progression of the disease to AML and explained these findings with an inefficient immune surveillance of transforming myelodysplastic syndrome clones by NK cells of individuals with a low number of activating KIR genes.46 However, they also found the same haplotype A distribution when comparing patients with de novo AML vs healthy individuals. Published data suggesting contradictory roles of KIR A/B haplotypes in the antileukemic activity of NK cells (or other KIR-expressing lymphocytes) might be related to the differences in relative population frequencies of KIR and KIR-ligand genes or are pointing at false positive results.
We observed a significantly higher frequency of C1/C1-positive individuals in the control group vs in patients with AML (39.4% vs 36.2%, respectively; adjusted P = .027), and observed a trend for patients with AML to expose C2/C2 more often than for the healthy individuals (14.1% vs 12.4%; adjusted P = .112) (supplemental Table 3; supplemental Figure 2). Furthermore, in our study, individuals homozygous for Bw4-80-I were more in the patient group (3.8% vs 2.5%; adjusted P = .007) (supplemental Table 3; supplemental Figure 3). NK cells expressing the activating KIR2DS1 receptor become hyporesponsive in C2/C2 individuals,43 and HLA molecules bearing the Bw4-80-I motifs can be bound by inhibitory KIR3DL1 receptors. However, the presence or absence of KIR2DS1 or KIR3DL1 did not explain the 2 aforementioned effects pointing at mechanisms other than KIR/KIR-ligand interactions.
Another possible explanation for the revealed associations with certain HLA types might be the peptide–major histocompatibility complex–dependent surveillance by T cells. One could speculate on the role of early mutations in the evolution of AML giving rise to immunogenic neoantigenic peptides derived from mutated proteins. Such peptides may be presented only by some HLA types, possibly belonging all to a putatively favorable (eg, C1) or unfavorable (eg, C2 or Bw4) HLA class I KIR-ligand group. Also, NK cells can recognize HLA molecules in a peptide-dependent manner, as described for the framework KIR gene KIR3DL2 binding to HLA-A3 and HLA-A11.47 For the latter 2 HLA alleles, a trend for a reduced frequency among adult patients with acute lymphoblastic leukemia was found by Deng et al.45 In the presented study, frequencies of HLA-A11 or HLA-A3/A11 did not differ when comparing patients with AML vs control subjects.
Probably the most plausible approach to classify individuals based on KIR gene information is to combine it with the KIR-ligand HLA genotype. Single KIR/KIR-ligand combinations have been addressed in the context of HCT. According to a study from Venstrom et al, KIR2DS1+ donors mediated a reduced relapse rate for patients with C1+ but not for those with C2/C2 with AML.23 We challenged this model in the very different immunological context of development of de novo AML but could not detect different frequencies of the respective groups among patients and controls, neither did we find differences when classifying according to a model proposed by Boudreau et al, focusing on different alleles of KIR3DL1/KIR3DS1 and their ligands Bw4 80-I or Bw4 80-T for grouping.22 More comprehensive models have been developed, taking even more inhibitory and/or activating KIRs and their cognate ligands into account. Such scores were predictive for the clinical outcome of infections with HIV, hepatitis C virus, and human T-lymphotropic virus19 or of unrelated HCTs among patients with AML or various hematologic malignancies.35,36 However, when we applied the published scores for grouping, we did not detect strong significant differences suggesting an effect on the generation of AML. For a low (≤1) inhibitory count and a low (≤1.75) inhibitory score based on the criteria given in the study by Boelen et al,19 a tendency to mediate protection from developing de novo AML was found, not supporting the theory in the original paper, which states that functional engagement of iKIRs enhances immune responses and improves clinical outcomes. Our data suggest that a high inhibitory burden might hamper successful immune surveillance of (pre)leukemic clones instead. This small effect could also be an incidental finding or point at different mechanisms underlying immune responses to viral infections and leukemogenesis.
Our study provides evidence that the individual KIR genotype does not constitute a risk factor for developing AML. These results suggest that steady-state KIR-mediated NK cell immunity does not define susceptibility for AML or that NK immunity is disarmed as leukemic clones grow. It is worth mentioning that specific NK cell subsets appear after cytomegalovirus (CMV) infection, which show phenotypic characteristics including NKG2C and KIR expression apart from playing a role in the prevention of AML relapse after HCT.48 Also, some evidence for an oncoprotective effect of CMV infection was shown for certain hematologic malignancies in a nontransplant setting.49 To elucidate the putative underlying immunologic mechanisms of CMV-associated and KIR-mediated anticancer, activity is warranted and might lead to new models of NK cell function in leukemia. Finally, it is reasonable to analyze the individual repertoires of KIR-expressing NK cell subsets and KIR expression levels in large flowcytometric studies to uncover their possibly decisive role for KIR-mediated endogenous immune surveillance of developing AML.
Acknowledgments
The authors thank Nicole Heymann, Heike Uhlemann, Dorothea Luise Weber, Anja Schieferdecker, and Marie Helbig for technical assistance; Michael Kramer for sample identification; and the staff of Collaborative Biobank, Study Alliance Leukemia Biobank, and DKMS Life Science Lab.
Authorship
Contribution: J. Schetelig, A.H.S., F.H., and H.B. designed research; F.H. and E.R.-B. prepared samples; C.M. and G.S. analyzed and interpreted raw sequencing data; B.F. and H.B. performed statistical analysis; F.H., B.F., and J. Schetelig wrote a first version of the manuscript; U.V.S., J. Sauter, F.S., M.J.M., M.v.B., and V.L. discussed the results and commented on the manuscript; and C.R., H.A., C.T., K.S.-E., C.M.-T., S.W.K., S.K., M.K., M.H., H.S., A.N., and M.B. contributed samples and scientific input during data analysis and discussion.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Falk Heidenreich, Clinical Trials Unit, DKMS gGmbH, Augsburger Straße 3, 01309 Dresden, Germany; e-mail: heidenreich@dkms.de.
References
Author notes
Sequencing data are provided with supplemental Table 2.
Data are available on request from the corresponding author, Falk Heidenreich (heidenreich@dkms.de). Requests will be checked carefully and data will be made available by the authors without undue reservation.
The full-text version of this article contains a data supplement.