Abstract
Immune thrombocytopenia (ITP) is the consequence of a complex, still incompletely understood immunological dysregulation. Proposed mechanisms include autoantibody-induced platelet destruction, impaired platelet production as well as abnormalities in T-cell immunity, such as T helper cells (Th1) polarization, a high proportion of Th17 cells, and a reduced number of regulatory T cells. Although the etiology of ITP is incompletely understood and considered multifactorial in most cases, genetic variants are thought to play a key role in susceptibility to ITP, especially in persistent or chronic ITP. Efforts are currently underway to uncover possible predisposing genetic factors for the development of ITP. Single-nucleotide polymorphisms and copy number variations have been identified in several immune-related genes, such as cytokine genes, Fcγ receptor genes or T-cell costimulation genes, and have been associated with patients’ susceptibility to ITP. However, because of the clinical heterogeneity and low incidence of ITP it remains challenging to perform genetic analyses with sufficiently large sample size within informative patient populations, highlighting the need for collection of well-annotated biomaterials in clinical trials or registry projects. Another significant challenge is to go beyond performing association studies alone and to establish genotype-phenotype associations, thus proving causality between a genetic alteration and ITP pathogenesis. This review summarizes our current knowledge on genetic alterations identified as potential predisposing factors for the development of ITP in adults, thereby addressing signaling pathways considered critical for ITP pathogenesis.
Introduction
Immune thrombocytopenia (ITP) is an immune-mediated disorder in which defects in immune self-tolerance lead to accelerated platelet destruction and impaired platelet production, both resulting in a thrombocytopenic state with variable bleeding symptoms.1 Once considered idiopathic, ITP is now seen as a condition with a complex autoimmune pathogenesis characterized by multiple processes of immune system dysregulation.2 The formation of antiplatelet autoantibodies is considered to be the pivotal mechanism of platelet destruction together with impaired megakaryopoiesis and thrombopoiesis in the bone marrow.3,4 More recently, T-cell–mediated processes have been shown to play an equally crucial role in the development and perpetuation of ITP.5-7 Several mechanisms, such as the disturbed balance of T helper cells (Th1/Th2),8,9 a decreased number and dysfunction of regulatory T cells,10,11 and cytotoxic T lymphocyte–mediated platelet destruction12,13 have been identified. Moreover, it is increasingly recognized that platelets are not only targets of autoinflammatory processes but might directly play an active role in ITP onset and progression via their role as both immune sensing and immune effector cells.14,15
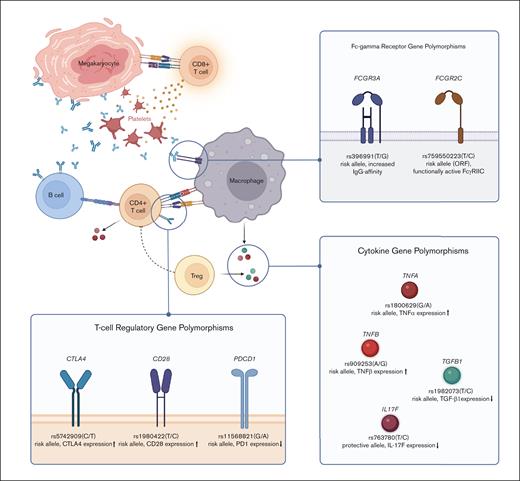
Despite the advances in understanding the pathogenesis of ITP, the principle immune mechanism that initiates ITP has not yet been identified. ITP is considered to be a multifactorial disease caused, and sustained by, interactions between environmental as well as genetic factors.16-19 Although the contribution of individual genetic factors, and their impact on the emergence and course of ITP are still largely unknown, efforts are underway to uncover potential host genetic factors associated with the development of ITP. Discovering causative genes or pathways is crucial for identifying populations that are at risk and for developing new therapeutic strategies that may prevent chronicity. Single-nucleotide polymorphisms (SNPs) and copy number variations (CNVs), the most common patterns of genetic variants, have been identified in various immunity-related genes, such as cytokine genes,20-23 Fcγ receptor (FcγR) genes,24-26 or T-cell costimulatory genes,27,28 and have been linked to patients’ susceptibility to develop ITP (Figure 1).
Selection of genetic polymorphisms associated with ITP susceptibility. Schematic illustration of the major signaling pathways in ITP pathophysiology and genetic polymorphisms identified in associated genes. The featured polymorphisms represent a selection of SNPs whose significant influence on ITP susceptibility and/or severity has been confirmed in several independent studies. Minor alleles and their effect on protein expression/function are written in bold. Figure created with BioRender.com.
Selection of genetic polymorphisms associated with ITP susceptibility. Schematic illustration of the major signaling pathways in ITP pathophysiology and genetic polymorphisms identified in associated genes. The featured polymorphisms represent a selection of SNPs whose significant influence on ITP susceptibility and/or severity has been confirmed in several independent studies. Minor alleles and their effect on protein expression/function are written in bold. Figure created with BioRender.com.
Although familial ITP is a very rare disease, only 2% of ITP cases reported to be familial,29 there is also some anecdotal evidence for a hereditary component in ITP development. Besides descriptive reports of ITP in monozygotic twins or families with multiple affected members30, recent family-based linkage studies used high-throughput sequencing methods that enabled the identification of germ line mutations in genes such as IKZF131 or TNFRSF13B32 as predisposing factors for familial ITP. The phenomenon of hereditary ITP has to be clearly separated from defined genetic disorders associated with thrombocytopenia, including common variable immunodeficiency, myosin heavy-chain 9–related disorders, autoimmune lymphoproliferative syndrome related to defects in the FAS pathway, and Wiskott-Aldrich syndrome/X-linked thrombocytopenia.
Although the clinical definition of ITP is similar between children and adults, there appear to be several profound differences in disease development.33 Childhood ITP is usually associated with a viral infection and tends to be self-limiting, with ∼80% of cases resolving spontaneously within 6 months. The transient nature of viral-mediated ITP in children is well established and was demonstrated to result from the occurrence of molecular mimicry between viral antigens and host platelet proteins.34 In contrast, adult ITP is rarely, or at least less evidently, triggered by infection and frequently develops into a chronic disease in which spontaneous resolution is uncommon, pointing to profound immunological differences between ITP in children and adults. Although the clinical condition in adolescents and adults with chronic ITP (cITP) appears similar, it remains questionable whether pediatric and adult cases can simply be pooled together for the purpose of genetic analysis. To our knowledge, the first review of potential predisposing genetic factors for pediatric ITP was published by Bergman et al in 2010.35 To account for potential immunological differences, this review will focus on genetic studies of adult ITP only. This review summarizes the current evidence on genetic alterations identified as predisposing factors for the development of ITP and highlights the associated signaling pathways considered crucial to the pathogenesis of ITP.
Literature search
A systematic search of PubMed/Medline (performed in September 2022) was conducted to identify appropriate studies. The following keyword combinations were used: ([ITP] OR [immune thrombocytopenia] OR [immune thrombocytopenic purpura] OR [idiopathic thrombocytopenic purpura] AND [polymorphism∗], [mutation∗], [gene∗], [variant∗], [gwas], [snp], and [molecular]. In addition, the reference lists of all retrieved articles were reviewed to identify additional sources. All studies published in English between 2000 and 2022 were individually assessed. Studies were excluded if 1 of the following conditions was fulfilled: (1) not related to ITP; (2) inappropriate publication type, including reviews, case reports, letters to the editors, correspondences, and abstracts; (3) insufficient data; or (4) exclusively dealing with pediatric ITP. A total of 36 publications were included.
Polymorphisms in cytokine and cytokine receptor genes
As a typical autoimmune disease, ITP is associated with the dysregulation of cytokine production and activity. In patients with cITP, several studies have shown a distinct Th1-polarized cytokine profile36,37 and a significantly higher Th1:Th2 ratio.9 Thus, Th1 polarization appears to be a central mechanism in the pathogenesis of ITP. In this context, the association between cytokine gene polymorphisms and ITP susceptibility has been extensively studied.
One of the main cytokines used to distinguish Th1 from other CD4+ subsets is interferon gamma (IFN-γ), which is secreted mainly by Th1 and natural killer cells and exerts proinflammatory activities.38 Among patients with cITP, several investigations have shown IFN-γ to be upregulated at both the messenger RNA (mRNA) and protein levels.8,9,39 In line with these findings and as further confirmation of a disturbed Th1:Th2 ratio, it was reported that serum levels of interleukin-4 (IL-4) are decreased in patients with cITP.39,40
The polymorphism rs2430561 of the IFN-γ gene (IFNG), which is located in the nuclear factor κB binding site of the IFN-γ promotor, has been reported to affect IFN-γ production, with the homozygous minor T/T genotype being associated with higher production of IFN-γ than with the A/A genotype.41 Several studies have already shown an association between the proinflammatory, high expression T/T genotype and the occurrence of various autoimmune diseases.42 In contrast, conflicting data have been reported in ITP. Although Pehlivan et al reported a significantly higher frequency of the T/T genotype in a cohort of Turkish patients with cITP compared with healthy controls,43 Chen et al observed no significant association between the IFNG rs2430561 T allele and ITP risk in a cohort of Chinese patients with cITP.44 A possible source of inconsistency between results in this case may be the study of diverse ethnic groups, because SNP distribution can vary greatly among different ethnicities.
The influence of different functional Th1/Th2 cytokine and cytokine receptor polymorphisms was further investigated by Takahashi et al.45 Although their data showed no significant association of the IFNG rs2430561 non-A/A genotype with ITP susceptibility, patients with the minor T allele or homozygous T/T genotype had more severe thrombocytopenia and a significantly higher Th1:Th2 ratio.45 Takahashi et al further demonstrated that patients with cITP with the mutant C/C genotype of IL-4 rs2243250, which reportedly is associated with lower IL-4 levels,46 had a higher number of treatment regimens than patients who did not have the C/C genotype. From these data, one might speculate that the IL-4 rs2243250 C/C genotype contributes to an impaired Th1/Th2 ratio, which in turn appears to lead to a more refractory disease course.
Satoh et al observed that the frequency of the mutant G/G genotype of tumor necrosis factor β polymorphism rs909253 was significantly higher in Japanese patients with ITP compared with that in healthy controls.20 The potential pathogenetic significance of this SNP was further supported by the group’s finding that frequencies of circulating antiglycoprotein IIb/IIIa antibody–producing B cells were significantly higher in patients with ITP with the homozygous mutant G/G genotype than in patients with the G/A or A/A genotypes. Taken together, these study results indicate that proinflammatory genotypes of IFN-γ pathway components contribute to a higher Th1:Th2 ratio in patients with ITP and may thus increase the likelihood and/or severity of ITP.
In addition to Th1-driven mechanisms, it is increasingly recognized that a specific lineage of effector T cells, that is, Th17 cells, plays an equally important role in the development of ITP.44,47 Th17 cells play a central role in mucosal and epithelial host defense against extracellular pathogens, whereas abnormal activation has been linked to the pathogenesis of several autoimmune diseases.48 There is more and more evidence that the circulating number of Th17 cells is significantly higher in patients with ITP than that in healthy controls.49-51 Furthermore, Hu et al found that the frequency of another subset of IL-17–producing CD8+ cells, the so called Tc17 cells, was significantly increased in patients with newly diagnosed ITP compared with that in patients who already had achieved a complete remission.52 Considering the key role of Th17/Tc17 in immunity, the effector cytokine IL-17 is thought to play a crucial role in autoimmune disease in general and also in the pathogenesis of ITP.53 Within all known members of the IL-17 family, IL-17F is best characterized and considered to be primarily responsible for the activity of Th17 cells.54-56 Saitoh et al examined an association between the IL-17F gene (IL-17F) SNP rs763780 and susceptibility to ITP.57 Functional in vitro analyses have previously shown that IL-17 expression and activity may be downregulated in carriers of the minor C allele.55 Saitoh et al found a significantly lower frequency of the minor C allele and C/C genotype in patients with cITP than that in healthy controls, suggesting that the IL-17F rs763780 C allele might be a protective factor for cITP development. Furthermore, patients with the homozygous T/T genotype showed a more severe thrombocytopenia at diagnosis than in patients with the T/C genotype. These results are consistent with studies performed by Liu et al and Li et al in Chinese Han populations, both showing that the frequency of the IL-17F rs763780 C allele was significantly decreased in patients with ITP.23,58 Tolba et al further highlighted the role of IL-17 rs763780 and ITP susceptibility in a cohort of Egyptian patients with ITP (pediatric and adult). In this study, the rare homozygous C/C genotype was exclusively detected in the control group. The minor C allele was also significantly more prevalent in the control group, again suggesting that the IL-17 rs763780 C allele is inversely associated with development of ITP and might have a protective effect.59
In addition to the aforementioned cytokines, polymorphisms in genes coding for several other cytokines or cytokine receptors have been investigated in adult ITP, for example, IL-1B, IL-10, IL-18, or IL-23R (summary in Table 1), for most of which associations remain uncertain or controversial and further studies are needed to elucidate their potential role in disease onset and treatment response.
Studies reporting genetic polymorphisms in immunoregulatory genes in patients with ITP
| Publication . | Genetic polymorphisms . | Cohort . | ||
|---|---|---|---|---|
| Ethnicity . | ITP type . | Patient numbers (patients with ITP; controls) . | ||
| Cytokine and cytokine receptor gene polymorphisms | ||||
| Satoh et al20 | •TNFA (rs1800629 and rs1800629)•TNFB (rs909253)•IL-1B (rs16944 and rs1143634) | Asian (Japanese) | Adult-onset, primary, and chronic | 84; 56 |
| Chen et al44 | •IFNG (rs2430561)•IL-4 (IVS3) | Asian (Chinese) | Adult- and childhood-onset, acute, persistent, and chronic | 196; 128 |
| Saitoh et al57 | •IL-17F (rs763780) | Asian (Japanese) | Adult-onset and chronic | 102; 188 |
| Pehlivan et al43 | •TNFA (rs1800629)•TGFB1 (rs1982073 and rs1800471)•IL-10 (rs1800896, rs1800871, and rs1800872)•IL-6 (rs1800796)•IL-1RA (VNTR)•IFNG (rs2430561) | Caucasian (Turkish) | Adult-onset and chronic | 71; 71 |
| Sarpatvari et al115 | •IL-10 (rs1800871 and rs1800872)•TNFA (rs1800629)•TGFB1 (rs4803457)•IL-1A (rs1800587)•IL-4RA (rs1801275) | Caucasian | Adult-onset and primary | 206; 618 |
| Zhan et al116 | •IL-23R (rs10889677, rs1884444, rs7517847, and rs11209032) | Asian (Chinese) | Adult-onset, primary, and acute | 75; 81 |
| Li et al117 | •IL-18 (rs187238 and rs1946518) | Asian (Chinese) | Adult- and childhood-onset, primary, and chronic | 181; 163 |
| Li et al118 | •IL-17F (rs763780) | Asian (Chinese) | Adult- and childhood-onset, primary, and chronic | 165; 149 |
| El Ghannam et al119 | •IL-10 (rs1800896, rs1800871, and rs1800872) | African (Egyptian) | Adult-onset and chronic | 62; 73 |
| Pavkovic et al87 | •IL-1B (rs16944)•TNFB (rs909253)•TNFA (rs1800629) | Caucasian (Macedonian) | Adult-onset and primary | 125; 120 |
| Liu et al23 | •IL-17A (rs2275913)•IL-17F (rs763780) | Asian (Chinese) | Adult-onset, primary, and chronic | 146; 137 |
| Yadav et al120 | •TNFA (rs1800629)•TNFB (rs909253) | Asian (Indian) | Adult-onset and primary | 80; 100 |
| Takahashi et al45 | •IFNG (rs2430561)•IFNGR (rs1327474)•IL-4 (rs2243250)•IL-4RA (rs1801275) | Asian (Japanese) | Adult-onset and chronic | 126; 202 |
| Yadav et al121 | •IL-1B (rs1143627 and rs16944)•IL-1RA (VNTR) | Asian (Indian) | Adult-onset and primary | 118; 100 |
| Li et al58 | •IL-2 (rs6822844) | Asian (Chinese) | Adult-onset and primary | 312; 154 |
| Yu et al98 | •IL-18 (rs1946518)•IL-1B (rs16944) | Asian (Chinese) | Adult- and childhood-onset and primary | 403; 336 |
| Tolba et al59 | •IL-17F (rs763780) | African (Egyptian) | Adult- and childhood-onset and primary | 105; 112 |
| Nomura et al122 | •TNFA (rs1800629)•TGFB1 (rs1982073)•IL-10 (rs1800896, rs1800871, and rs1800872)•IFNG (rs2430561) | Asian (Japanese) | Adult-onset | 153; 70 |
| T-cell regulatory gene polymorphisms | ||||
| Du et al99 | •TIM3 (rs10053538, rs10515746, and rs1036199) | Asian (Chinese) | Adult- and childhood-onset, primary, acute, and chronic | 187; 123 |
| Li et al58 | •DNAM1 (rs763361) | Asian (Chinese) | Adult-onset and primary | 312; 154 |
| Kasamatsu et al28 | •PDCD1 (rs36084323 and rs11568821),•CTLA4 (rs733618, rs11571316, rs231775, and rs3087243) | Asian (Japanese) | Adult-onset and chronic | 119; 223 |
| Abdelghafar et al79 | •CD40 (rs1883832 and rs4810485) | African (Egyptian) | Adult-onset and primary | 101; 97 |
| Wang et al70 | •TIM3 (rs10515746)•CD28 (rs1980422)•TNFSF4 (rs2205960)•CTLA4 (rs231779)•PDCD1 (rs36084323)•ICOS (rs6726035)•DNAM1 (rs763361)•LAG3 (rs870849) | Asian (Chinese) | Adult-onset and primary | 307; 295 |
| Chen et al68 | •CTLA4 (rs11571315, rs733618, rs4553808, rs11571316, rs62182595, rs16840252, rs5742909, rs231775, and rs3087243)•CD28 (rs1879877, rs3181096, rs3181097, rs3181098, rs56228674, rs3116496, and rs3116496) | Asian (Taiwanese) | Adult-onset, primary, and secondary | 32; 30 |
| Ellithy et al80 | •CD40 (rs1883832 and rs4810485) | African (Egyptian) | Adult-onset and chronic | 50; 50 |
| FcγR gene polymorphisms | ||||
| Fujimoto et al25 | •FCGR2A (rs1801274)•FCGR3A (rs396991) | Asian (Japanese) | Adult-onset, primary, and chronic | 104; 59 |
| Breunis et al26 | •FCGR2A (rs1801274)•FCGR2B (rs1050501)•FCGR3A (rs396991)•FCGR3B HNA1a/HNA1b/HNA1c•FCGR2C (rs759550223) | Caucasian | Adult- and childhood-onset | 116; 100 |
| Xu et al123 | •FCGR2B (rs1050501) | Asian (Chinese) | Adult- and childhood-onset, acute, and chronic | 280; 243 |
| Xu et al124 | •FCGR2A (rs1801274)•FCGR3A (rs396991) | Caucasian, Asian, and African | Adult- and childhood-onset | 741;1092 (meta-analysis) |
| Audia et al88 | •FCGR2A (rs1801274)•FCGR2B (rs1050501)•FCGR3A (rs396991)•FCGR3B HNA1a/HNA1b/HNA1c•FCGR2C (rs759550223) | Caucasian | Adult-onset and primary | 216; 88 |
| Pavkovic et al87 | •FCGR2A (rs1801274)•FCGR3A (rs396991) | Caucasian (Macedonian) | Adult-onset, primary, and chronic | 125; 120 |
| Li et al89 | •FCGR2A (rs1801274)•FCGR2B (rs1050501)•FCGR3A (rs396991) | Caucasian and Asian | Adult- and childhood-onset | 1200; 1723 (meta-analysis) |
| Polymorphisms in miscellaneous genes | ||||
| Pehlivan et al43 | •GP1A (rs1126643)•GP2 (VNTR)•GP3A (rs4918)•MBL2 (rs1800450) | Caucasian (Turkish) | Adult-onset and chronic | 71; 71 |
| Basciano et al125 | •TUBB1 (rs6070697) | — | Adult-onset and chronic | 191; 361 |
| Ge et al94 | •PTPN22 (rs2488457) | Asian (Chinese) | Adult- and childhood-onset, primary, acute, and chronic | 191; 216 |
| Li et al58 | •CD24 (rs52812045)•FCRL3 (rs945635, rs7528684, rs3761959, and rs11264799)•IRF5 (rs2280714, rs2004640, and rs10954213)•ITGAM (rs1143679)•NLRP3 (rs4353135, rs35829419, and rs10754558)•CARD8 (rs2043211)•PTPN22 (rs33996649 and rs1310182)•SH2B3 (rs3184504)•STAT4 (rs10181656 and rs7574869)•TNFAIP3 (rs10499194, rs2230926, rs5029939, and rs6920220)•TRAF1 (rs10818488) | Asian (Chinese) | Adult-onset and primary | 312; 154 |
| Yu et al98 | •NLRP3 (rs35829419)•CARD8 (rs2043211)•NFKB1 (rs28362491) | Asian (Chinese) | Adult- and childhood-onset and primary | 403; 336 |
| Hassan et al96 | •TLR9 (rs352140) | African (Egyptian) | Adult-onset and primary | 40; 40 |
| Zhang et al97 | •FOXP3 (rs5902434, rs3761548, and rs2232365) | Asian (Chinese) | Adult- and childhood-onset, primary, and chronic | 329; 279 |
| Tian et al126 | •PTPN22 (rs2476601 and rs2488457) | Caucasian, Asian, and African | Adult- and childhood-onset, and primary | 932; 2112 (meta-analysis) |
| GWASs | ||||
| Xu et al101 | •GBE1/LINC02027 (rs117503120)•TIMP3/SYN3 (rs5998634)•TENM4 (rs4483616)•OSBPL6 (rs16866133) | Asian (Chinese) | Adult-onset and primary | 200; 200 |
| Publication . | Genetic polymorphisms . | Cohort . | ||
|---|---|---|---|---|
| Ethnicity . | ITP type . | Patient numbers (patients with ITP; controls) . | ||
| Cytokine and cytokine receptor gene polymorphisms | ||||
| Satoh et al20 | •TNFA (rs1800629 and rs1800629)•TNFB (rs909253)•IL-1B (rs16944 and rs1143634) | Asian (Japanese) | Adult-onset, primary, and chronic | 84; 56 |
| Chen et al44 | •IFNG (rs2430561)•IL-4 (IVS3) | Asian (Chinese) | Adult- and childhood-onset, acute, persistent, and chronic | 196; 128 |
| Saitoh et al57 | •IL-17F (rs763780) | Asian (Japanese) | Adult-onset and chronic | 102; 188 |
| Pehlivan et al43 | •TNFA (rs1800629)•TGFB1 (rs1982073 and rs1800471)•IL-10 (rs1800896, rs1800871, and rs1800872)•IL-6 (rs1800796)•IL-1RA (VNTR)•IFNG (rs2430561) | Caucasian (Turkish) | Adult-onset and chronic | 71; 71 |
| Sarpatvari et al115 | •IL-10 (rs1800871 and rs1800872)•TNFA (rs1800629)•TGFB1 (rs4803457)•IL-1A (rs1800587)•IL-4RA (rs1801275) | Caucasian | Adult-onset and primary | 206; 618 |
| Zhan et al116 | •IL-23R (rs10889677, rs1884444, rs7517847, and rs11209032) | Asian (Chinese) | Adult-onset, primary, and acute | 75; 81 |
| Li et al117 | •IL-18 (rs187238 and rs1946518) | Asian (Chinese) | Adult- and childhood-onset, primary, and chronic | 181; 163 |
| Li et al118 | •IL-17F (rs763780) | Asian (Chinese) | Adult- and childhood-onset, primary, and chronic | 165; 149 |
| El Ghannam et al119 | •IL-10 (rs1800896, rs1800871, and rs1800872) | African (Egyptian) | Adult-onset and chronic | 62; 73 |
| Pavkovic et al87 | •IL-1B (rs16944)•TNFB (rs909253)•TNFA (rs1800629) | Caucasian (Macedonian) | Adult-onset and primary | 125; 120 |
| Liu et al23 | •IL-17A (rs2275913)•IL-17F (rs763780) | Asian (Chinese) | Adult-onset, primary, and chronic | 146; 137 |
| Yadav et al120 | •TNFA (rs1800629)•TNFB (rs909253) | Asian (Indian) | Adult-onset and primary | 80; 100 |
| Takahashi et al45 | •IFNG (rs2430561)•IFNGR (rs1327474)•IL-4 (rs2243250)•IL-4RA (rs1801275) | Asian (Japanese) | Adult-onset and chronic | 126; 202 |
| Yadav et al121 | •IL-1B (rs1143627 and rs16944)•IL-1RA (VNTR) | Asian (Indian) | Adult-onset and primary | 118; 100 |
| Li et al58 | •IL-2 (rs6822844) | Asian (Chinese) | Adult-onset and primary | 312; 154 |
| Yu et al98 | •IL-18 (rs1946518)•IL-1B (rs16944) | Asian (Chinese) | Adult- and childhood-onset and primary | 403; 336 |
| Tolba et al59 | •IL-17F (rs763780) | African (Egyptian) | Adult- and childhood-onset and primary | 105; 112 |
| Nomura et al122 | •TNFA (rs1800629)•TGFB1 (rs1982073)•IL-10 (rs1800896, rs1800871, and rs1800872)•IFNG (rs2430561) | Asian (Japanese) | Adult-onset | 153; 70 |
| T-cell regulatory gene polymorphisms | ||||
| Du et al99 | •TIM3 (rs10053538, rs10515746, and rs1036199) | Asian (Chinese) | Adult- and childhood-onset, primary, acute, and chronic | 187; 123 |
| Li et al58 | •DNAM1 (rs763361) | Asian (Chinese) | Adult-onset and primary | 312; 154 |
| Kasamatsu et al28 | •PDCD1 (rs36084323 and rs11568821),•CTLA4 (rs733618, rs11571316, rs231775, and rs3087243) | Asian (Japanese) | Adult-onset and chronic | 119; 223 |
| Abdelghafar et al79 | •CD40 (rs1883832 and rs4810485) | African (Egyptian) | Adult-onset and primary | 101; 97 |
| Wang et al70 | •TIM3 (rs10515746)•CD28 (rs1980422)•TNFSF4 (rs2205960)•CTLA4 (rs231779)•PDCD1 (rs36084323)•ICOS (rs6726035)•DNAM1 (rs763361)•LAG3 (rs870849) | Asian (Chinese) | Adult-onset and primary | 307; 295 |
| Chen et al68 | •CTLA4 (rs11571315, rs733618, rs4553808, rs11571316, rs62182595, rs16840252, rs5742909, rs231775, and rs3087243)•CD28 (rs1879877, rs3181096, rs3181097, rs3181098, rs56228674, rs3116496, and rs3116496) | Asian (Taiwanese) | Adult-onset, primary, and secondary | 32; 30 |
| Ellithy et al80 | •CD40 (rs1883832 and rs4810485) | African (Egyptian) | Adult-onset and chronic | 50; 50 |
| FcγR gene polymorphisms | ||||
| Fujimoto et al25 | •FCGR2A (rs1801274)•FCGR3A (rs396991) | Asian (Japanese) | Adult-onset, primary, and chronic | 104; 59 |
| Breunis et al26 | •FCGR2A (rs1801274)•FCGR2B (rs1050501)•FCGR3A (rs396991)•FCGR3B HNA1a/HNA1b/HNA1c•FCGR2C (rs759550223) | Caucasian | Adult- and childhood-onset | 116; 100 |
| Xu et al123 | •FCGR2B (rs1050501) | Asian (Chinese) | Adult- and childhood-onset, acute, and chronic | 280; 243 |
| Xu et al124 | •FCGR2A (rs1801274)•FCGR3A (rs396991) | Caucasian, Asian, and African | Adult- and childhood-onset | 741;1092 (meta-analysis) |
| Audia et al88 | •FCGR2A (rs1801274)•FCGR2B (rs1050501)•FCGR3A (rs396991)•FCGR3B HNA1a/HNA1b/HNA1c•FCGR2C (rs759550223) | Caucasian | Adult-onset and primary | 216; 88 |
| Pavkovic et al87 | •FCGR2A (rs1801274)•FCGR3A (rs396991) | Caucasian (Macedonian) | Adult-onset, primary, and chronic | 125; 120 |
| Li et al89 | •FCGR2A (rs1801274)•FCGR2B (rs1050501)•FCGR3A (rs396991) | Caucasian and Asian | Adult- and childhood-onset | 1200; 1723 (meta-analysis) |
| Polymorphisms in miscellaneous genes | ||||
| Pehlivan et al43 | •GP1A (rs1126643)•GP2 (VNTR)•GP3A (rs4918)•MBL2 (rs1800450) | Caucasian (Turkish) | Adult-onset and chronic | 71; 71 |
| Basciano et al125 | •TUBB1 (rs6070697) | — | Adult-onset and chronic | 191; 361 |
| Ge et al94 | •PTPN22 (rs2488457) | Asian (Chinese) | Adult- and childhood-onset, primary, acute, and chronic | 191; 216 |
| Li et al58 | •CD24 (rs52812045)•FCRL3 (rs945635, rs7528684, rs3761959, and rs11264799)•IRF5 (rs2280714, rs2004640, and rs10954213)•ITGAM (rs1143679)•NLRP3 (rs4353135, rs35829419, and rs10754558)•CARD8 (rs2043211)•PTPN22 (rs33996649 and rs1310182)•SH2B3 (rs3184504)•STAT4 (rs10181656 and rs7574869)•TNFAIP3 (rs10499194, rs2230926, rs5029939, and rs6920220)•TRAF1 (rs10818488) | Asian (Chinese) | Adult-onset and primary | 312; 154 |
| Yu et al98 | •NLRP3 (rs35829419)•CARD8 (rs2043211)•NFKB1 (rs28362491) | Asian (Chinese) | Adult- and childhood-onset and primary | 403; 336 |
| Hassan et al96 | •TLR9 (rs352140) | African (Egyptian) | Adult-onset and primary | 40; 40 |
| Zhang et al97 | •FOXP3 (rs5902434, rs3761548, and rs2232365) | Asian (Chinese) | Adult- and childhood-onset, primary, and chronic | 329; 279 |
| Tian et al126 | •PTPN22 (rs2476601 and rs2488457) | Caucasian, Asian, and African | Adult- and childhood-onset, and primary | 932; 2112 (meta-analysis) |
| GWASs | ||||
| Xu et al101 | •GBE1/LINC02027 (rs117503120)•TIMP3/SYN3 (rs5998634)•TENM4 (rs4483616)•OSBPL6 (rs16866133) | Asian (Chinese) | Adult-onset and primary | 200; 200 |
Genetic polymorphisms significantly associated with ITP susceptibility and/or severity are written in bold.
CARD8; caspase recruitment domain family member 8; FCGR2A, Fcγ receptor IIa; FCGR2B, Fcγ receptor IIb; FCGR2C, Fcγ receptor IIc; FCGR3A, Fcγ receptor IIIa; FCGR3B, Fcγ receptor IIIb; FCRL3, Fcγ receptor like 3; FOXP3, Forkhead box P3; GBE1, 1,4-α-glucan branching enzyme 1; GP1A, glycoprotein Ia; GP2, glycoprotein II; GP3A, glycoprotein IIIa; ICOS, inducible T-cell costimulator; IFNGR, interferon-γ receptor 1; IL-1A, interleukin-1α; IL-1RA, IL-1 receptor antagonist; IL-1B, IL-1β; IL-4RA, IL-4 receptor subunit α; IRF5, interferon regulatory factor 5; ITGAM, integrin subunit α M; LAG3, lymphocyte-activation gene 3; LINC02027, long intergenic nonprotein coding RNA 2027; MBL2, mannose binding lectin 2; NFκB1, nuclear factor κB subunit 1; NLRP3, NLR family pyrin domain containing 3; OSBPL6, oxysterol binding protein like 6; PDCD1, programmed cell death protein 1; PTPN22, protein tyrosine phosphatase nonreceptor type 22; SH2B3, SH2B adapter protein 1; STAT4, signal transducer and activator of transcription 4; SYN3, synapsin III; TENM4, teneurin transmembrane protein 4; TGFB1, transforming growth factor β1; TIM3 T-cell immunoglobulin and mucin domain containing 3; TIMP3, tissue inhibitor of metalloproteinase 3; TNFA, tumor necrosis factor α; TNFAIP3, TNF-α–induced protein 3; TLR9, Toll-like receptor 9; TNFB, tumor necrosis factor β; TNFRSF5, TNF receptor superfamily member 5; TNFSF4, TNF superfamily member 4; TRAF1, TNF receptor–associated factor 1; TUBB1, tubulin β1 class VI.
Polymorphisms in genes regulating T-cell activity
Immune checkpoints, including costimulatory and coinhibitory signaling pathways, are among the central regulatory mechanisms of T-cell–mediated immune responses and pivotal for maintaining self-tolerance.60-62 The costimulatory molecules of T cells consist of CD28, inducible costimulatory (ICOS), TNF ligand superfamily member 4 (TNFSF4), and DNAX accessory molecule 1 (DNAM1); the coinhibitory molecules include T-cell immunoglobulin and mucin domain containing 3 (TIM3), cytotoxic T lymphocyte–associated protein 4 (CTLA-4), programmed cell death protein 1 (PDCD1), and lymphocyte-activation 3 (LAG3).63-67 Of these, CD28 and CTLA-4 are considered of critical importance and have been extensively studied.
Chen et al recently investigated the prevalence of different SNPs in the promoter region of CTLA4 and CD28 in patients with ITP and the effects of these SNPs on gene expression.68 Genotype frequencies of CTLA4 rs11571315 and rs5742909 differed significantly between ITP cases and healthy controls, yet rs5742909 reached statistical significance only in the group of patients with secondary ITP. Investigation of the biological effects of these SNPs revealed that the minor T allele of rs5742909 was associated with an 80% decrease in transcriptional activity of a CLTA-4 reporter. Based on these results, the authors speculate that polymorphisms in the promoter region of CTLA4 contribute to T-cell overactivation due to the reduced expression of the coinhibitory molecule CTLA-4 and, thus, contribute to the pathogenesis of ITP. However, these findings might be biased because of the inclusion of patients with secondary ITP, which is mainly caused by other autoimmune diseases. It must be considered that polymorphisms found in the CTLA4 promoter are primarily related to the underlying autoimmune disease and not necessarily causal for the development of a thrombocytopenic state.
In addition to their potential impact on susceptibility, there is some evidence suggesting that polymorphisms in immune checkpoint–related genes may influence the clinical features of ITP. In a Japanese cohort, Kasamatsu et al found that the CTLA4 rs11571316 G allele and rs3087243 G allele, both of which correlate with a low expression of CTLA-4, were associated with a significantly higher than normal frequency of bleeding symptoms and lower platelet counts at diagnosis.28 These results are consistent with the findings of Guo et al, showing that the CD28/CTLA-4 ratio of circulating T cells in patients with cITP was inversely correlated with platelet counts.69 In addition to CTLA4 and CD28, Wang et al investigated the contribution of SNPs in several other immune checkpoint–related genes (ie, ICOS, PDCD1, TNFSF4, TIM3, DNAM1, and LAG3) to ITP susceptibility.70 Their results indicated that the CD28 rs1980422 C allele was associated with a significantly increased risk of primary ITP. At the functional level, the authors further demonstrated that CD28 expression was significantly increased at both the mRNA and protein levels in patients with the C/T genotype compared with those with the T/T genotype. Furthermore, Wang et al found that the PDCD1 rs36084323 T allele and the DNAM1 rs763361 T allele to be associated with ITP severity and corticosteroid-resistance, respectively.70
In addition to the molecules mentioned earlier, the CD40/CD40 ligand (CD40L) axis is thought to play a critical role in the pathogenesis of various autoimmune diseases, including ITP.71,72 The interaction of CD40L on activated T cells with its receptor CD40 on B cells is essential for B-cell proliferation, isotype switch, and generation on memory B cells. In ITP, perturbations in the CD40/CD40L interaction have been demonstrated by significantly elevated levels of CD40L on the platelet surface, potent to induce proliferation of B cells in spatial proximity to potential autoantigens, such as platelet glycoproteins.73-75 Hence, interruption of the CD40-CD40L interaction via anti-CD40L monoclonal antibodies has been considered a possible therapeutic strategy in patients with refractory ITP.76 Regarding genetic alterations in this pathway, it has been proposed that the CD40 polymorphism rs1883832 affects the interaction between CD40 and CD40L via altered expression of CD40 on B cells. Currently, it has not been definitively clarified whether this is due to an increased expression of CD40 and, thus, increased stimulation of autoreactive B cells77 or a decreased expression of CD40 leading to the decreased formation of CD8+ regulatory T cells, which are the central mediators of immune tolerance.78 AbdelGhafar et al evaluated the impact of the CD40 polymorphism rs1883832 on the risk of developing ITP in 101 Egyptian patients and found that carriers of the mutant T/T genotype had a significantly higher risk of developing ITP, indicating an association of CD40 polymorphisms and ITP susceptibility.79 Similar findings were obtained by Ellithy et al, who found that carriers of combined gene polymorphisms CD40 rs1883832 and CD40 rs4810485 have a higher risk of developing cITP.80
Taken together, these results clearly point to an important role of costimulatory molecules in ITP, but further exploration of the genetic diversity in these pathways is urgently needed to unravel their potential association and, more importantly, causality in the context of ITP development.
FcγR polymorphisms
FcγR-mediated phagocytosis of antibody-sensitized platelets is the primary pathogenic mechanism by which autoantibodies induce thrombocytopenia.81 The central role of FcγR in ITP is underscored by the fact that intravenous immunoglobulin treatment and splenectomy (removal of the platelet-destructing organ) are proven treatment options.82 In FcγR biology, balanced coexpression of both activating (FcγRI, FcγRIIa, FcγRIIc, FcγRIIIa, and FcγRIIIb) and inhibitory receptors (FcγRIIb) is considered to be crucial for preventing autoreactivity.83 Genetic polymorphisms affecting FcγR expression or immunoglobulin G subclass binding were found primarily in genes encoding activating FcγRs, such as FCGR2A,84FCGR3A,85 and FCGR3B.86
In ITP, Fujimoto et al investigated the role of FcγR polymorphisms for disease susceptibility by assessing the frequencies of FCGR2A rs1801274 and FCGR3A rs396991 in 104 Japanese patients with cITP.25 Results revealed a significantly lower frequency of the FCGR3A rs396991 wild-type T/T genotype in patients with ITP, whereas the frequency of FCGR2A rs1801274 genotypes did not differ between patients and controls. Furthermore, patients with the mutant G/G genotype of FCGR3A rs396991 showed significantly higher complete remission rates after treatment with corticosteroids or other immunosuppressive agents than patients with the G/T or T/T genotypes, suggesting that FCGR3A polymorphisms might affect both ITP susceptibility and treatment response.
One of the largest studies published by Pavkovic et al investigated the genotype distributions and allele frequencies for FCGR2A rs1801274 and FCGR3A rs396991 in 125 Macedonian patients with cITP.87 In accordance with Fujimoto et al, frequency of the mutant G allele of FCGR3A rs396991 differed significantly between patients with ITP and controls as well as between patients with steroid-responsive ITP and those with steroid-unresponsive ITP. Overrepresentation of the FCGR3A rs396991 G allele was also observed in a study by Audia et al who examined FcγR polymorphisms in human splenic macrophages from 24 patients with primary ITP who had undergone splenectomy in comparison with such polymorphisms in posttraumatic splenectomy control specimens.88
A unique investigation of the impact of genetic variation of the FcγR gene cluster on predisposition to ITP was performed by Breunis et al. The group developed a multiplex ligation-dependent probe amplification assay that allows for the simultaneous identification of both SNPs and CNVs in FCGR2A, FCGR2B, FCGR2C, FCGR3A, and FCGR3B.26 In a study comparing the frequency of FcγR gene variations in patients with ITP (children and adults) and healthy controls, results confirmed the previously observed overrepresentation of the FCGR3A rs396991 G allele. Moreover, they identified a significant overrepresentation of a FCGR2C open reading frame allele in patients with ITP. This activating open reading frame allele is determined by a SNP in exon 3 (rs759550223) of the pseudogene FCGR2C and was shown to result in the expression of a functionally active FcγRIIc.
Recently, a large comprehensive meta-analysis on FcγR polymorphisms and ITP has been published, combining the results of the aforementioned studies as well as various publications on pediatric ITP. The pooled analyses of pediatric and adult patients with ITP performed by Li et al showed a significant association of the FCGR3A rs396991 G allele with a higher risk for both childhood-onset and adult ITP, suggesting that this polymorphism can indeed be considered a predisposing factor for ITP development.89
In summary, polymorphisms in FcγR genes have been shown to contribute to genetic susceptibility to ITP, although the exact nature of their contribution remains poorly understood. Moreover, rather than asingle genetic variant, multiple SNPs and CNVs within the FcγR cluster collectively cause a change in the balance between activating and inhibitory receptor signals.
In addition to improving our understanding of predisposing factors for ITP emergence, exploring polymorphisms in FcγR genes could help to identify potential predictors of treatment response. Because blockage of FcγRs has been proven useful in the therapeutic management of ITP,90 a difference in FcγR expression due to genetic variants could have a pivotal influence on the efficacy of such treatment modalities. Studies on the impact of FcγR polymorphisms on clinical factors or therapeutic responses in ITP, however, are scarce. A study conducted by Cooper et al provided evidence that the rs1801274 and rs396991 polymorphisms of FCGR2A and FCGR3A, respectively, may affect cytokine and platelet responses after anti-D treatment.91 Ellity et al explored the influence of the aforementioned gene polymorphisms on the response to rituximab in patients with cITP and observed an association of the wild-type C allele of FCGR2A rs1801274 with a better response to rituximab.92 Although available data are limited, the influence of FcγR polymorphisms on therapy response is an intriguing research area and could potentially influence treatment decisions in the future.
Genetic variants in miscellaneous genes
Based on previous research findings and a pathogenetic understanding of other autoimmune diseases, several other immune-related genes have been investigated for a possible association with an individual’s susceptibility to ITP. Li et al performed a comprehensive analysis on the contribution of 25 SNPs in 13 inflammation-related genes to disease susceptibility and treatment response in a cohort of 312 Chinese patients with primary ITP.58 The selected SNPs were chosen based on their clear association with autoimmunity in previously published work. Data analysis showed a significant difference in the frequency of rs10499194 in the TNF-α–induced protein 3 gene (TNFAIP3) between patients with ITP and healthy controls, suggesting that the minor T allele of this SNP may be a protective factor for the development of ITP. The group further demonstrated a genotype-phenotype correlation; individuals with the C/T genotype on the TNFAIP3 rs10499194 locus had higher TNFAIP3 mRNA expression compared with patient with the wild-type C/C genotype. Interestingly, the rs10499194 T allele was also associated with corticosteroid sensitivity.
Part of the analysis by Li et al as well as several other working groups was analyzing a potential relationship between ITP development and polymorphisms in PTPN22, a gene encoding the protein tyrosine phosphatase nonreceptor type 22, which is critically involved in the regulation of signal transduction via T-cell receptors.93 Available research on this topic has shown conflicting results, with some studies insufficiently powered to show significant results.58,94,95 A recent meta-analysis conducted by Tian et al, which included 10 studies with a total of 932 cases, showed that the SNPs rs2476601 and rs2488457 in PTPN22 may contribute to genetic susceptibility to primary ITP.
Additional candidate genes, which are under investigation for a potential impact on ITP risk, include genes encoding components of the inflammasome,96 regulators of regulatory T-cell function,97 Toll-like receptors,98 or T-cell immunoglobulin and mucin domain family proteins.99
The majority of studies focused on identifying genetic factors associated with susceptibility for, or persistence of, ITP, thereby obtaining further insights into the pathophysiology of the disease. However, in ITP, there is an equally compelling need for predictive biomarkers that allow for better prediction and monitoring of treatment response. An interesting contribution in this respect has been made by Basciano et al, who investigated a SNP in β1-tubulin chain gene (TUBB1) as a response biomarker in patients with ITP. β1-tubulin is primarily responsible for the assembly of platelet microtubules and thus essential for platelet production and function. The study showed that allele frequencies of TUBB1 rs6070697 did not differ between 191 patients with ITP and 361 healthy controls, indicating that the SNP does not play a direct etiologic role in ITP. However, patients with ITP with the minor A allele (both heterozygous and homozygous) received significantly more lines of therapy and had a significantly higher risk of failure with immunomodulatory therapies, particularly rituximab. Although the detailed pathological mechanisms remain to be elucidated in this case, these data again highlight the potential of genetic variants to serve as biomarkers in ITP.
Genome Wide Association Studies
As previously demonstrated, several susceptibility loci for ITP have been identified using traditional candidate gene approaches, including direct sequencing and polymerase chain reaction restriction fragment length polymorphism analysis. These approaches are largely limited by their reliance on prior knowledge of the functional aspects of potential candidate genes, which can lead to selection bias. In contrast, genome-wide association studies (GWASs), which take a more holistic approach, are a powerful tool in identifying novel gene variants in complex diseases.100
So far, the number of GWASs in ITP is very limited. Xu et al performed a pooling GWAS involving 200 ITP cases and 200 controls from a Chinese Han population.101 Via SNP microarrays and pools analysis, their study revealed SNPs in 4 novel loci (glycogen branching enzyme, tissue inhibitor of metalloproteinase 3 [TIMP3], teneurin transmembrane protein 4, and RNA binding motif protein 45) to be strongly associated with ITP. For some of these genes, literature research already reveals a potential pathogenetic link to ITP. TIMP3 for example is an inhibiting matrix metalloproteinase protein,102 which has been shown to have a statistically positive correlation with IL-4 levels and platelet count but a negative correlation with IFN-γ levels in patients with ITP, suggesting that this protein may contribute to Th1-polarization.103 In addition, this study found further evidence for the pathogenetic relevance of the JAK-STAT pathway in ITP, which is reportedly involved in the etiological mechanism of pediatric ITP104 and plays a key role in the pharmacological mode of action of the thrombopoietin receptor agonist eltrombopag.105 The results of this study show that GWASs can provide a powerful stimulus for in-depth assessment of previously understudied genetic loci and may potentially reveal novel pathways linked to the etiology of ITP.
Current challenges and future directions
Although the etiology of ITP is incompletely understood and considered multifactorial in most cases, genetic variants are thought to play a pivotal role in the susceptibility to ITP, especially in cases of persistent or cITP. Accordingly, to date, several studies have focused on the association between host genetic factors in immunologically relevant genes and ITP.
There is considerable evidence in the literature that polymorphisms in immunity-associated genes contribute to the susceptibility of ITP and, probably, also to the progression and chronicity of the disease. The most convincing data on the matter have been obtained for genes such as IFN-y, IL-4, and IL-17 as well as for individual members of the FcγR family. In addition to these genes, there is a large data set on genetic alterations in ITP that is difficult to interpret and lacks the scope for generalization. Because of the low incidence of ITP and the fact that sporadic cases are highly heterogeneous, designing genetic analyses with sufficiently large sample sizes and informative patient populations remains challenging. Most reported studies deal with small sample numbers, limiting the power of these analyses to clearly identify certain genetic associations. Therefore, it is critical to first perform calculations on the minimum sample size, depending primarily on whether it is an analysis of candidate genes with a limited number of independent tests or a GWAS scenario, and on the allele frequency and expected relative risk of the genetic variant under investigation.106 As important as calculating the required sample size is defining the phenotype of interest as precisely and specifically as possible, taking into account disease etiology, disease duration, and patient age. Nonspecific case definition will increase both the genetic and environmental heterogeneity in underlying causal factors and can therefore decrease the power of detection of an effect. Furthermore, replication of a study will be difficult if the phenotype has not been adequately defined. In ITP, care should be taken when selecting cases for genetic studies to avoid mixing primary and secondary ITP as well as acute and chronic disease, at a minimum. For such a rare disease as ITP, these considerations clearly highlight the importance of collecting well-annotated biomaterial in clinical trials or registry projects.
Association studies with candidate genes are a valuable approach to elucidate the genetic background of a complex disorder such as ITP. Such studies, which typically include patient cases and healthy age- and sex-matched controls, can be performed in a relatively simple and economic manner and allow the identification of genes that have small effects. Unfortunately, this approach is hampered by its reliance on prior knowledge of possible candidate genes, which makes selection bias unavoidable.
In ITP, a large number of candidate gene association studies shows inconsistent results or insufficient statistical significance. The differences in these results can be explained mainly by small sample size, biological heterogeneity, and selection of different subgroups of patients, as well as statistical bias because of multiple testing procedures.
In contrast, GWASs usually proceed without any prior assumptions regarding the functional significance of the investigated genes. The identified risk loci often implicate genes of unknown function or of previously unsuspected relevance, thereby motivating experimental follow-up and potentially leading to the discovery of novel disease mechanisms.100 However, there is concern that most of the identified genotype-phenotype associations reflect variants and genes with no direct biological relevance to the underlying disease. However, with a rare disease such as ITP, the major challenge is again to achieve a sufficient sample size to conduct a GWAS with reliable and conclusive results. The smaller the effect and the lower the prevalence of any given polymorphism, the larger a population needs to be for the gene to be discovered. To complicate matters, the distribution of SNPs can vary greatly between different ethnic groups, making it essential to have controls that match the ethnic background of the study population. Overrepresentation of different ethnic groups in previous analyses complicates the accurate interpretation of the existing genetic data on ITP because there may be a difference in ITP pathophysiology between Western and Asian populations. For example, an association between HLA and ITP has been found in Asian populations,107,108 whereas every attempt to assess this in Western populations has failed. Therefore, it remains to be verified whether the results obtained in predominantly Asian populations can be generalized to other ethnic groups.
To add further value to data collected on the genetic background of ITP, research agendas should go beyond conducting association studies alone and should aim to demonstrate clear genotype-phenotype associations. After all, studies on various other autoimmune diseases have shown similar associations with polymorphisms as those observed in ITP,42,109-111 which raises the question of whether any discovered polymorphism is actually causative for ITP or potentially related to autoimmunity in general. Future studies need to investigate whether there is a direct causal relationship between different polymorphisms and ITP and to determine the relevance of any alteration for the defining thrombocytopenic state. The most valuable approach to achieve this ambitious goal is certainly the use of well-designed murine models, which have already contributed substantially to the understanding of the basic genetic and proteomic mechanisms of ITP.112-114 In the future, the generation of knockin mouse models that allow detailed functional analysis of the effects of genetic alterations on molecular and cellular processes in vivo may help elucidate the impact of genetic predisposition in ITP.
The complex etiology of ITP and its diverse clinical presentation most likely results from an equally heterogeneous genetic background. Many different SNPs may affect the response to different environmental factors and precipitating events. This makes the identification of a definite driver mutation highly unlikely. Consequently, there are currently no genetic markers that are individually sufficiently powerful to provide a causal explanation for the development of ITP. However, there is no question that further genetic analyses will lead the way in deciphering the pathogenic mechanisms of ITP and identifying potential biomarkers or treatment targets required for stratified and targeted therapies.
Authorship
Contribution: J.-A.G. wrote the manuscript; J.M.M., M.B., A.M., and K.T.-G. read and edited the manuscript; and all authors contributed to the article and approved the submitted version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Julia-Annabell Georgi, Medizinische Klinik und Poliklinik I, Universitätsklinikum Carl Gustav Carus der Technischen Universität, Fetscherstraße 74, 01307 Dresden, Germany; e-mail: julia-annabell.georgi@uniklinikum-dresden.de.
References
Author notes
Data are available on request from the corresponding author, Julia-Annabell Georgi (julia-annabell.georgi@uniklinikum-dresden.de). No individual patient data will be shared.