Key Points
We developed a tool to facilitate the selection of HLA-DPB1-mismatched unrelated transplantation donors.
HLA-DPB1 sequence features in exons 2 and 3 provide optimal information regarding risky mismatches.
Abstract
HLA-DP is a classic transplantation antigen that mediates alloreactivity through T-cell epitope (TCE) diversity and expression levels. A current challenge is to integrate these functional features into the prospective selection of unrelated donor candidates for transplantation. Genetically, HLA-DPB1 exon 2 defines the permissive and nonpermissive TCE groups, and exons 2 and 3 (in linkage with rs9277534) indicate low- and high-expression allotypes. In this study, we analyzed 356 272 exon 2-exon 3–phased sequences from individuals across 5 self-identified race and ethnicity categories: White, Hispanic, Asian or Pacific Islander, Black or African American, and American Indian or Alaskan Native. This sequence data set revealed the complex relationship between TCE and expression models and the importance of exon 3 sequence data. We also studied archived donor search lists for 2545 patients who underwent transplantation from an HLA-11/12 unrelated donor mismatched for a single HLA-DPB1 allele. Depending on the order in which the TCE and expression criteria were considered, some patients had different TCE- and expression-favorable donors. In addition, this data set revealed that many expression-favorable alternatives existed in the search lists. To improve the selection of candidate donors, we provide, disseminate, and automate our findings through our multifaceted tool called Expression of HLA-DP Assessment Tool, consisting of a public web application, Python package, and analysis pipeline.
Introduction
HLA matching between the patient and unrelated donor for hematopoietic cell transplantation is undertaken to lower the risk of adverse outcomes, including graft-versus-host disease (GVHD) and mortality. Typically, matching is first ensured for HLA-A, -B, -C, -DRB1, and -DQB1 before HLA-DPB1 matching is evaluated. In the case of HLA-DPB1, multiple models have emerged through retrospective studies validated across several large patient cohorts, demonstrating that some HLA-DPB1 mismatches are less risky (T-cell epitope [TCE]-permissive; low-expression) than others (TCE-nonpermissive; high-expression). These models are based on the categorization of HLA-DPB1 according to their assigned TCE group or HLA-DP expression level.
The TCE model depends on the T-cell recognition of epitopes in the peptide-binding region.1,2 Because of the role of this model in GVHD and mortality risk,3,4 researchers have made several adjustments over the decades, for example, the refinement of TCE group categories,5-7 analysis of amino acid variation,6,8,9 characterization of the immunopeptidome,10-12 and testing of directionality.13 To maintain compatibility with the existing National Marrow Donor Program (NMDP) infrastructure, we used the algorithm developed by Crivello et al6 that uses the immunogenicity model (with the most immunogenic TCE group as 1 and the least immunogenic TCE group as 3) and leverages functional distance calculations.8 The NMDP recommends this algorithm in its 2019 unrelated donor-matching guidelines,14 with further considerations15 for implementing TCE core alleles.5
The HLA-DP expression model describes the immunogenicity of low- or high-expression HLA-DP allotypes of the patient that can be recognized by the donor. This applies to single mismatches, in which low HLA-DP expression in the mismatched allotype of the patient denotes a favorable mismatch with a low risk of acute GVHD. Conversely, high HLA-DP expression mismatches are high-risk and unfavorable.16 HLA-DP expression is associated with the rs9277534 single nucleotide polymorphism (SNP) in the 3′ untranslated region (UTR), in which the rs9277534A allele correlates with low expression, and the rs9277534G allele with high expression.16 rs9277534 is in strong positive linkage disequilibrium (LD) with a series of identified SNPs within exon 3.17 In the absence of 3′ UTR sequence data, exon-3 variation serves as a proxy for expression levels with a high degree of accuracy for White European populations.17 Because of the lack of experimental data on HLA-DP expression and TCE alloreactivity, complete sequence data are most desirable to pinpoint associations and understand patterns in more diverse populations.
When combining both models, some transplantation pairs are concordant with TCE-permissive + low-expression and TCE-nonpermissive + high-expression mismatches. Other pairs are discordant with TCE-nonpermissive + low-expression or TCE-permissive + high-expression mismatches. Prior studies have reported 62% to 76% concordance18-20 when both models were applicable with single HLA-DPB1 mismatches. Notably, the TCE model accommodates multiple mismatches, whereas the expression model is informative for single mismatches. In the absence of a fully matched donor, when selecting a donor using both models to lower the risk of acute GVHD, studies recommend identifying a low-expression single mismatch followed by a TCE-permissive mismatch.16,19,21,22
In this study, we leverage available large population collections with extensive sequencing data together with prior models and outcome studies to (1) provide further insight into correlations between commonly sequenced regions and more definitive HLA-DPB1 biomarker associations, (2) develop an integrated tool called ExPAT (Expression of HLA-DP Assessment Tool) to apply both TCE and expression models to HLA-DPB1 donor alleles and donor-recipient mismatches, and (3) show how the application of these models can impact donor selection. Our tool enables the use of maximal complex sequence information for all models in an accessible manner for researchers, physicians, and transplant providers.
Methods
Study population
We studied 3 data sets, as described below. For all 3, we extracted the NMDP self-identified race and ethnicity (SIRE) categories linked to US census categories23 (eg, AFA [Black or African American], API [Asian or Pacific Islander], CAU [White], HIS [Hispanic or Latin American], and NAM [American Indian or Alaskan Native]) from individuals in the NMDP database. We excluded individuals whose responses did not match these categories (eg, unknown, multiple, other, and multirace).
Be The Match registry donors (data set 1)
For data set 1, we obtained 356 188 HLA-DPB1 sequences containing exons 2 and 3 from 178 096 Be The Match Registry donors categorized between 2016 and 2020. NMDP-contracted laboratories determined these sequences using whole-genome sequencing. These sequences were reported, stored in, and extracted using the histoimmunogenetics markup language format developed by the NMDP Bioinformatics research group.24 ExPAT annotated each sequence for TCE and expression features.
Be The Match stem cell sources containing rs9277534 (data set 2)
For data set 2, we extracted 550 descriptive sequences for exon 3 and rs9277534 in the 3′UTR of HLA-DPB1. NMDP-contracted laboratories generated these sequences using single-molecule real-time sequencing developed by Pacific Biosciences. These sequences originated from recruited donors (65.1%) and cord blood units during the confirmatory typing stage (34.9%). Using phased data, data set 2 confirmed the physical linkage between exon 3 and rs9277534 in the 3′UTR. Similar to data set 1, ExPAT annotated the sequences extracted from histoimmunogenetics markup language files.
Retrospective donor-recipient transplantation pairs (data set 3)
For data set 3, we collated 5032 patients who had previously received transplantations between 2008 and 2021 from HLA-A, -B, -C, -DRB1, and -DQB1–matched (HLA-10/10) unrelated donors stored in an NMDP database called Search Archive 1.0. Of these patients, 2545 (50.6%) had a single HLA-DPB1 mismatch (HLA-11/12). Additionally, we extracted the archived donor search lists at the time of the search as calculated by the HapLogic search algorithm.25 ExPAT annotated the expression and TCE matching categories for the patients and each of their potential donors. Analysis scripts determined whether better expressions and TCE alternatives were present in the archived search lists.
Development of the package, microservice, UI of ExPAT, and analysis scripts
We developed ExPAT to automate the annotation of TCE and expression features of alleles, genotypes, and recipient-donor (mis)matches. ExPAT consists of a Python 3 package, Flask-RESTPlus microservice, and an Angular 14 user interface (UI). The code and instructions for using these components are available at https://github.com/nmdp-bioinformatics/dpb1-expression. Additionally, the GitHub subdirectory analysis houses Python scripts that drive the analyses of the described data sets 1, 2, and 3.
Assignment of HLA-DPB1 TCE groups and expression levels inferred from HLA nomenclature
The Immuno Polymorphism Database-international ImMunoGeneTics information system/Human Leukocyte Antigen (IPD-IMGT/HLA) database and GitHub repository (https://github.com/ANHIG/IMGTHLA)26 contain all the reference data necessary to assign known or unknown TCE groups and expression levels inferred from exon 3 sequence information for all alleles. Although the database has directly annotated TCE groups in a dedicated file, additional parsing is required to extract the 7 expression-linked exon 3 SNPs. Furthermore, the IMGT database reports only unambiguous typing information. As a solution, ExPAT automates exon 3 parsing and ambiguous typing handling via its Python Flask-RESTful microservice algorithm.
ExPAT accepts a variety of HLA typing that have allelic (World Health Organization–reported alleles) or nonallelic typing. The current HLA nomenclature organizes alleles with known, unambiguous sequences via 4 colon-delimited hierarchical fields: first (distinguishing different allele families or groups), second (HLA proteins), third (alleles with unique silent coding-region substitutions), and fourth field (alleles with unique noncoding region substitutions).27
Allelic typing is associated with an unambiguous DNA sequence. When submitted, an allele’s sequence is parsed for expression-linked exon 3 SNPs (rs1126537, rs1126541, rs1042187, rs1042212, rs1042331, rs104335, and rs1071597)28 to infer high or low expression. If the SNPs do not align completely (100%) with the ACCACTC or GTTGTCT haplotypes, then ExPAT computes the edit distances and reports the most similar haplotype. If exon 3 is not available, then the expression remains unknown (supplemental Figure 1).
Conversely, nonallelic typing can be used to identify multiple DNA sequences. This group includes 2-field HLA proteins, 1/3/4-field nonallelic typing, G groups (alleles with the same antigen recognition domain nucleotide sequence encoded by exon 2 for class II HLA molecules), P groups (alleles with the same amino acid sequence encoded by exon 2), Multiple allele codes (https://hml.nmdp.org/MacUI/), genotype list strings,29 etc. ExPAT expands nonallelic typing to all possible alleles and then determines the most prevalent expression-linked exon 3 SNPs, if available. As a minimum for HLA matching, the G and P groups included only exon 2 alleles. Because these groups omit exon 3, their expression levels can be ambiguous. When ambiguities in exon 3 SNPs occurred in nonallelic typing, the Common, Intermediate, and Well Documented 3.030 statuses for the possible high-resolution alleles were used to estimate the most likely inferred expression level. To report ambiguous typing expression levels, the ExPAT Python package uses prefixes. Tilde (∼) symbols indicate a less common minor allele, and question mark (?) prefixes denote that there are 2 or more possible alleles with the same Common, Intermediate, and Well Documented status but different inferred expression levels. Supplemental Figure 1 shows an example of an ambiguous assignment.
Another dimension of these assignments was whether they were experimentally confirmed or predicted. Experimentally, for example, Petersdorf et al directly determined the HLA-DP expression in 81 rs9277534-homozygous (AA/GG) individuals using a quantitative polymerase chain reaction assay.16 For TCE groups, alloreactive T-cell cross-reactivity assays defined TCE group assignments.6 If no experimental confirmation was available, then ExPAT relied on predictions. In the expression model, exon 3 SNPs helped predict expression levels. For TCE, predictions use functional distances of sequences compared with HLA-DPB1∗09:01 with specific amino acids in the antigen recognition domain.6 ExPAT reports all this information to ensure transparency to users.
Results
HLA-DPB1 TCE and expression variation
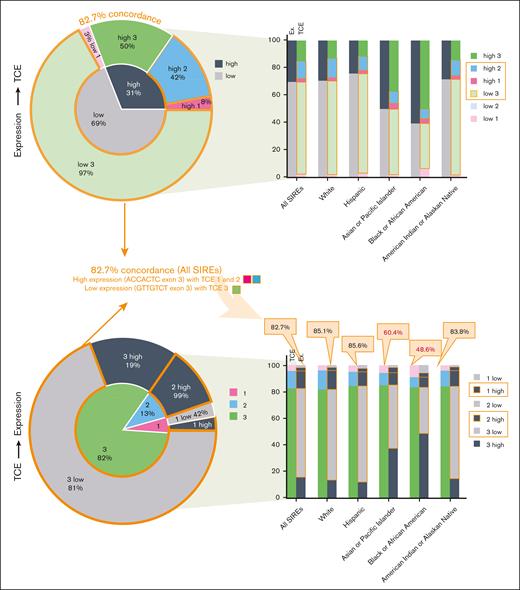
Previous studies on HLA-DPB1 expression-linked variation lacked sequences from ethnically diverse donors. This study contained 356 188 HLA-DPB1 sequences in data set 1 from 178 096 unrelated donors across 5 major SIRE categories: CAU (70.8%), HIS (20.3%), API (4.9%), AFA (3.6%), and NAM (0.3%). These sequences contained the phased exon 2 and exon 3 sequences. Exon 2 characterizes the TCE groups defined by Crivello et al.6 Exon 3 encodes a 7-SNP motif at a high LD with rs9277534 (Figure 1).
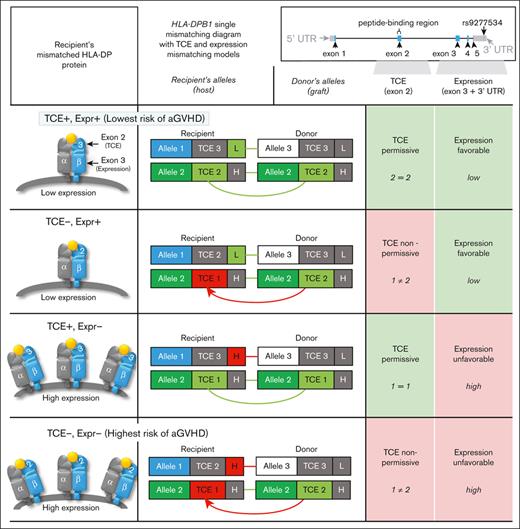
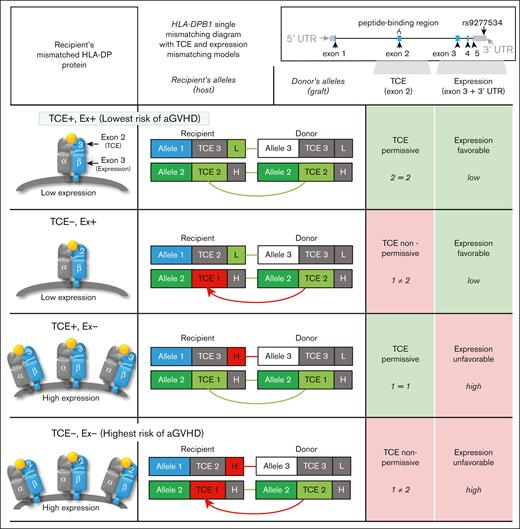
HLA-DPB1 TCE (exon 2–encoded) and HLA-DP expression (exon-3 and rs9277534-linked) (mis)matching model types. The immunogenicity TCE model involves the most immunogenic TCE groups in the recipient and donor (TCE 1 being the most immunogenic and TCE 3 being the least). Between these groups, matching is TCE permissive and mismatching is nonpermissive. Expression-favorable mismatching (applicable only to HLA-DPB1 single mismatches) involves the expression of the mismatched HLA-DP of the recipient inferred from a 7-SNP motif in exon 3 of HLA-DPB1. Low expression is more favorable than high expression.
HLA-DPB1 TCE (exon 2–encoded) and HLA-DP expression (exon-3 and rs9277534-linked) (mis)matching model types. The immunogenicity TCE model involves the most immunogenic TCE groups in the recipient and donor (TCE 1 being the most immunogenic and TCE 3 being the least). Between these groups, matching is TCE permissive and mismatching is nonpermissive. Expression-favorable mismatching (applicable only to HLA-DPB1 single mismatches) involves the expression of the mismatched HLA-DP of the recipient inferred from a 7-SNP motif in exon 3 of HLA-DPB1. Low expression is more favorable than high expression.
The expression of HLA-DP was inferred based on the 7-SNP exon 3 motif. Most of data set 1 (99.96%) contained 7 SNP exon 3 motifs as ACCACTC or GTTGTCT that are linked to high- or low-expression, respectively, as demonstrated by Schöne et al.17 To confirm the high LD rates between the exon 3 motif and rs9277534 in the 3′UTR, data set 2 consisted of 550 sequences with phased types from exon 3 to the 3′UTR. All these sequences confirmed the known ACCACTC-rs9277534G and GTTGTCT-rs9277534A linkages, which correlated with high- and low-expression, respectively (supplemental Table 1). Without this sequence information and using only HLA typing nomenclature, some ambiguities in HLA-DP expression assignment exist (supplemental Figures 2 and 3).
A comparison of the expression and TCE models illustrates the differing variations across SIRE categories within each model. Figure 2 displays the calculated allele frequencies for both the models. On comparing models, frequencies for exon 3–inferred expression levels appeared to vary more across SIRE categories than those for exon 2–characterized TCE groups. For instance, API and AFA SIRE categories have proportionally fewer low-expression alleles than other SIRE categories (Figure 2).
Frequencies of TCE and expression alleles in data set 1 containing 356 188 phased exon 2-exon 3 sequences across 5 SIRE categories. The top half consists of a sunburst chart (left) showing expression-to-TCE allele frequencies for all sequences and grouped stacked bar charts (right) that stratify frequency information into individual SIREs. The bottom half shows the same information but in reverse: from TCE to expression. Concordant sequences are defined as having TCEs 1 and 2 with high HLA-DP expression or TCE 3 with low expression. The concordant groups are outlined in orange. Each sequence links to an immunogenic TCE group based on version 2 assignments of IPD-IMGT/HLA. Exon 3 SNPs (rs1126537, rs1126541, rs1042187, rs1042212, rs1042331, rs104335, and rs1071597) in each sequence infer HLA-DP expression levels.
Frequencies of TCE and expression alleles in data set 1 containing 356 188 phased exon 2-exon 3 sequences across 5 SIRE categories. The top half consists of a sunburst chart (left) showing expression-to-TCE allele frequencies for all sequences and grouped stacked bar charts (right) that stratify frequency information into individual SIREs. The bottom half shows the same information but in reverse: from TCE to expression. Concordant sequences are defined as having TCEs 1 and 2 with high HLA-DP expression or TCE 3 with low expression. The concordant groups are outlined in orange. Each sequence links to an immunogenic TCE group based on version 2 assignments of IPD-IMGT/HLA. Exon 3 SNPs (rs1126537, rs1126541, rs1042187, rs1042212, rs1042331, rs104335, and rs1071597) in each sequence infer HLA-DP expression levels.
When TCE and expression models were combined, 82.7% of these allotypes fell into groupings in which high expression occurred in TCE 1 and 2 groups, and low expression occurred in TCE 3 group. Stratifications across SIREs revealed that API and AFA groups have more discordant groupings (Figure 2). We also performed the same analysis for core/noncore alleles5 (supplemental Figure 4). Despite the high concordance between both models, high variation in expression alleles across SIRE categories and limited overlap in certain SIREs can lead to downstream impacts on the hematopoietic stem cell trnsplantation donor selection.
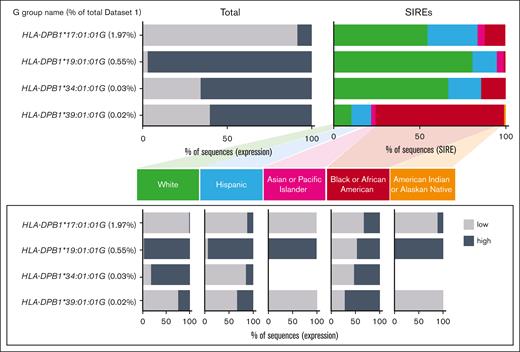
Imputation of HLA-DP expression: role of race and exon 3 sequence diversity
HLA-DPB1 exon 3 defines the β2 membrane-proximal domain and is not routinely typed. To gain information on exon 3 and better understand the discordance between TCE-expression (Figure 2), we determined specific exon-2 G groups that diverged into different exon-3 low/high-expression motifs. In data set 1, we identified several G groups that contained the same exon-2 sequence but differed in their exon-3 motifs. We set a >5% minor expression allele frequency criterion for motif admixtures within any population, in which the less common expression motif composed ∼5% to 50%, and the more common motif composed ∼50% to 95% of the sequences. Across the SIRE categories for these G groups, sequences in the AFA Americans demonstrated the highest diversity in exon 3. Conversely, sequences in API Americans demonstrated the least diversity in these same G groups (Figure 3).
The 4 exon-2-grouped sequences (G groups) in data set 1 of 356 188 phased exon 2-exon 3 sequences with an admixture of exon 3–low/high-expression motifs that contain ≥5% minor expression allele frequency within any of the SIRE categories. The ≥5% minor expression allele frequency occurs when the less common expression motif composes 5% to 50%, and the more common motif composes 50% to 95% of the sequences. These G groups represent 2.57% of data set 1.
The 4 exon-2-grouped sequences (G groups) in data set 1 of 356 188 phased exon 2-exon 3 sequences with an admixture of exon 3–low/high-expression motifs that contain ≥5% minor expression allele frequency within any of the SIRE categories. The ≥5% minor expression allele frequency occurs when the less common expression motif composes 5% to 50%, and the more common motif composes 50% to 95% of the sequences. These G groups represent 2.57% of data set 1.
TCE and expression: application to donor selection
To assess the ability of ExPAT to identify candidate donors for future patients using an expression-TCE algorithm, we studied a retrospective cohort of HLA-11/12 (HLA-A, -B, -C, -DRB1, and -DQB1 matched with HLA-DPB1 single mismatch) patient/donor transplantation pairs (data set 3). We filtered 2545 HLA-11/12 expression-applicable patients who underwent transplantationfrom 5032 HLA-10/10 patients who underwent transplantation (50.6%). Overall, 58.5% of the expression-relevant transplantations were concordant between the expression and TCE models. We further stratified this analysis for core/noncore TCE alleles, in which no expression-unfavorable mismatches had TCE core permissive mismatches (which involve the low expression HLA-DPB1∗02:01, 04:01, 04:02, and 23:01 alleles5; supplemental Figure 5).
Although the TCE model accommodates 1 or 2 HLA-DPB1 mismatches, the expression model applies to single HLA-DPB1 mismatches; particularly, for patients with 1 high- and 1 low-expression allotype, the expression model favors matching for the high-expression allotype to lower risks.2 In data set 3, ExPAT revealed that 43.1% of patients have an HL expression genotype. Within this subset of patients for whom the expression model is relevant, most (71.9%) had expression-unfavorable transplantations (TCE-nonpermissive + high-expression and TCE-permissive + high-expression; Figure 4B).
A multicomponent analysis of data set 3 examining TCE-expression concordance rates, distributions of TCE-expression mismatches and expression-favorable alternatives, and median HLA-11/12 (HLA-A, -B, -C, -DRB1, and -DQB1-matched 10/10 + HLA-DPB1 single mismatch) donor search list sizes. The match category shorthand includes Ex– (expression-unfavorable), Ex+ (expression-favorable), TCE+ (TCE-permissive), and TCE– (TCE-nonpermissive). (A) Cross tabulation of patients’ immunogenic TCE and expression (mis)match categories and concordance and discordance rates between both models. Concordance rates are the proportion of concordant matches (green and red) within all expression-relevant matches (total of Ex+ and Ex–). NA indicates matches that were not applicable to the expression model (patients with homozygous high [H] or low [L] expression genotypes: HH or LL). (B) Distribution of HLA-DPB1 match types (based on expression and TCE) for 2545 donor-recipient transplantations in data set 3 conducted from 2008 to 2021 for 9.2% HH, 43.1% HL, and 47.6% LL expression genotype patients, who were also stratifed based on individual SIRE categories. The expression-relevant information is outlined in gold. (C) Median list sizes of 11/12 donor search lists across SIRE categories.
A multicomponent analysis of data set 3 examining TCE-expression concordance rates, distributions of TCE-expression mismatches and expression-favorable alternatives, and median HLA-11/12 (HLA-A, -B, -C, -DRB1, and -DQB1-matched 10/10 + HLA-DPB1 single mismatch) donor search list sizes. The match category shorthand includes Ex– (expression-unfavorable), Ex+ (expression-favorable), TCE+ (TCE-permissive), and TCE– (TCE-nonpermissive). (A) Cross tabulation of patients’ immunogenic TCE and expression (mis)match categories and concordance and discordance rates between both models. Concordance rates are the proportion of concordant matches (green and red) within all expression-relevant matches (total of Ex+ and Ex–). NA indicates matches that were not applicable to the expression model (patients with homozygous high [H] or low [L] expression genotypes: HH or LL). (B) Distribution of HLA-DPB1 match types (based on expression and TCE) for 2545 donor-recipient transplantations in data set 3 conducted from 2008 to 2021 for 9.2% HH, 43.1% HL, and 47.6% LL expression genotype patients, who were also stratifed based on individual SIRE categories. The expression-relevant information is outlined in gold. (C) Median list sizes of 11/12 donor search lists across SIRE categories.
We also determined whether these expression-unfavorable transplantations had an expression-favorable donor available because data set 3 also contained alternative HLA-11/12 donor options (via HLA-DPB1 search typing) at the time of the search. Within this portion of expression-unfavorable transplantations, 56.0% had expression-favorable alternatives available (Figure 4B). These findings highlight the need for a tool to identify expression-favorable mismatches.
The availability of expression-favorable mismatches differed across the SIRE categories in data set 3. For these APIs and AFA patients, fewer expression-favorable alternatives were available (Figure 4B). Similarly, these 2 SIRE categories also had smaller median 11/12 donor list sizes (Figure 4C).
Emerging data from transplantation centers suggests that the order of candidate donor selection may be crucial for reducing the posttransplantation risks.22 Regardless of the order in which expression and TCE are applied, our data show that 2505 (98.4%) patients had the same final candidate donor choice (Figure 5B). In contrast, 40 (1.6%) patients had different donor choices when using the 2-step filtering algorithm. These patients did not have a donor concordantly permissive for both models but had options for both types of discordant donors in the search list: (1) favorable for expression but not TCE and (2) favorable for TCE but not expression (supplemental Figure 6B).
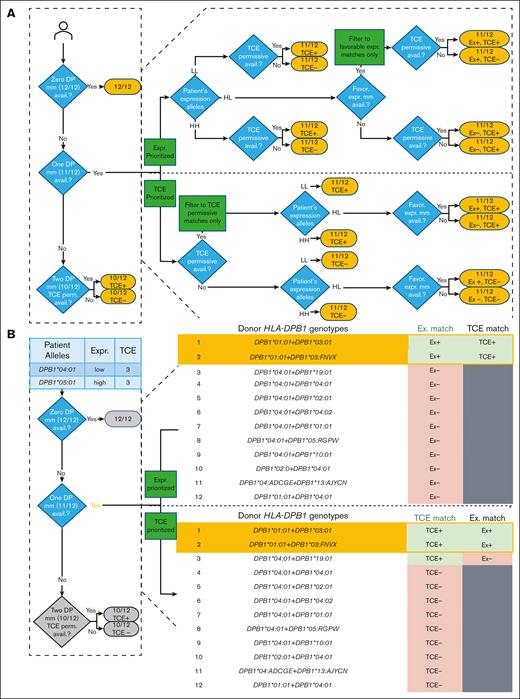
Flow diagram and example of how a combined TCE-expression algorithm can be used to identify an optimal HLA-11/12 donor (HLA-A, -B, -C, -DRB1, and -DQB1-matched 10/10 + HLA-DPB1 single mismatch) in data set 3. This data set involves 2545 HLA-11/12 expression-applicable patients who underwent transplantation filtered from 5032 HLA-10/10 (50.6%) patients who underwent transplantation. (A) Expression (Ex)-prioritized and TCE-prioritized routes for categorizing HLA-DPB1 match types between the recipient and donor. (B) An example search showing different final candidate HLA-11/12 search lists between expression-prioritized or TCE-prioritized applications. The example of search typing includes multiple allele codes that encode several possible alleles. Match category shorthand includes Ex– (expression-unfavorable), Ex+ (expression-favorable), TCE+ (TCE-permissive), and TCE– (TCE-nonpermissive). The golden boxes outline the final candidate donors after applying and filtering models. The grayed out boxes indicate that the donor did not pass the filtering step of the first model.
Flow diagram and example of how a combined TCE-expression algorithm can be used to identify an optimal HLA-11/12 donor (HLA-A, -B, -C, -DRB1, and -DQB1-matched 10/10 + HLA-DPB1 single mismatch) in data set 3. This data set involves 2545 HLA-11/12 expression-applicable patients who underwent transplantation filtered from 5032 HLA-10/10 (50.6%) patients who underwent transplantation. (A) Expression (Ex)-prioritized and TCE-prioritized routes for categorizing HLA-DPB1 match types between the recipient and donor. (B) An example search showing different final candidate HLA-11/12 search lists between expression-prioritized or TCE-prioritized applications. The example of search typing includes multiple allele codes that encode several possible alleles. Match category shorthand includes Ex– (expression-unfavorable), Ex+ (expression-favorable), TCE+ (TCE-permissive), and TCE– (TCE-nonpermissive). The golden boxes outline the final candidate donors after applying and filtering models. The grayed out boxes indicate that the donor did not pass the filtering step of the first model.
Automation of these findings via the HLA-DPB1 expression and TCE tool
ExPAT has multiple components available: a Python package/microservice (https://github.com/nmdp-bioinformatics/dpb1-expression), analysis scripts (https://github.com/nmdp-bioinformatics/dpb1-expression/tree/main/analysis), and a UI web application (https://dpb1-tce-expression.nmdp.org/). These presented findings were automated using these components and are available for public use. We provide several capabilities as outlined in Figure 6A.
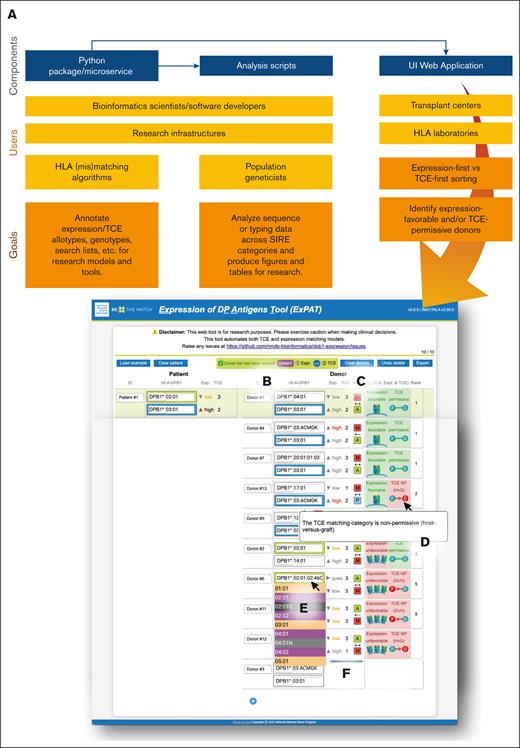
A diagram showing ExPAT’s utilities for several user types and a wide array of capabilities. (A) The UI (user interface) web application provides several functionalities via the immunogenic TCE model: (B) reversible expression and TCE donor sorting, (C) matching information (HapLogic grades and vector directionality), (D) allele expansion, (E) tooltips, (F) HLA typing autocompletion, and (G) real-time calculations.
A diagram showing ExPAT’s utilities for several user types and a wide array of capabilities. (A) The UI (user interface) web application provides several functionalities via the immunogenic TCE model: (B) reversible expression and TCE donor sorting, (C) matching information (HapLogic grades and vector directionality), (D) allele expansion, (E) tooltips, (F) HLA typing autocompletion, and (G) real-time calculations.
Figure 6 shows some of the capabilities of the UI. ExPAT provides TCE and expression assignments by comparing donor and patient HLA-DPB1 typing and suggests prioritization of favorable donors (TCE-permissive + low-expression mismatch) with customizable sorting (Figure 6B). ExPAT calculates HLA-matching information using HapLogic match grades25 and matching vector directionality (Figure 6C). Elements on the graphical UI contain tooltips to inform the user by expanding abbreviated terms and icons (Figure 6D) without presenting too much information upfront. The autocomplete functionality enables convenient and potential HLA typing inputs (Figure 6E). ExPAT also expanded the ambiguous alleles (supplemental Figure 7). HLA-matching (including TCE and expression) calculations were performed in real time (Figure 6F) to aid versatile clinical decision-making.
Discussion
Major advances in the immunobiology of HLA-DP in hematopoietic cell transplantation have been made over the last 2 decades, emphasizing the multifaceted pathways that define the immunogenicity of HLA-DP.1,16,31 The earliest models of HLA-DP alloreactivity were founded using T-cell epitopes as targets for host-versus-graft and graft-versus-host allorecognition.1,3 Many studies have confirmed the TCE model in independent transplantation cohorts, and the use of TCE for the selection of unrelated donors has been widely embraced to lower the risk of severe acute GVHD in unrelated donor HCT.3,32 More recently, 2 additional aspects of the function of HLA-DP have been uncovered. The Predicted Indirectly Recognizable HLA Epitopes model predicts polymorphic peptides processed from recipient-donor mismatched HLA molecules, for example, HLA-DP, and presented by mature HLA molecules on either recipient or donor for indirect allorecognition.31 Recently, a third model was developed based on the results of an major histocompatibility complex–wide SNP mapping study. In a discovery cohort of patients who received HLA-matched unrelated donor transplantations, the SNP rs2281389 was associated with the risk of acute GVHD, and the SNP association was replicated in an independent cohort.33 Fine-mapping of rs2281389 identified it to be in complete LD with HLA-DPB1 haplotypes inclusive of the 3′UTR region rs9277534 marker. Previous studies have identified the same regulatory region in the outcome of hepatitis B infection.34 The HLA-DPB1 regulatory haplotypes led to the third model of HLA-DP allorecognition, in which the level of expression of HLA-DP provides information on GVHD risk.16,19 Each of the 3 models describes unique features of HLA-DP molecules, but the structure of HLA-DP necessarily introduces an overlap between the epitopes that define peptide-binding and their linked regulatory variation.35 Studies have sought to bridge models for a unified approach to matching donors for transplantation.20,22 A retrospective analysis of a large cohort of HLA-matched and HLA-mismatched unrelated donor transplantations confirmed an association between high-expression single locus HLA-DPB1 mismatches and inferior transplantation outcome, and that in the setting of HLA-11/12 matching, the level of expression of the patient’s mismatched HLA-DP allotype provided information for donor selection.22 However, because the expression model is designed to interrogate single HLA-DP mismatches, when donors are mismatched for both HLA-DP allotypes, the (non) permissivity of TCE helps guide donor selection the best. This collective experience demonstrates that the application of epitope- and expression-based models for clinical decision-making depends on the patient’s unique HLA genotypes across the major histocompatibility complex and the pool of available unrelated donors. Hence, a preferred approach to lower risks for patients using the information on HLA-DP must be tailored to the patient’s unique HLA genetics.
In this study, we sought to demonstrate the feasibility of integrating TCE and expression into a tool for prospective donor selection. For a tool to be widely applicable to all patients and donors, it should be descriptive of the known HLA-DPB1 alleles for the sequence features that describe TCEs and expression levels. Because the TCE model is based on polymorphisms that define the peptide-binding domain, complete information on exon 2 at a minimum is required; however, exon 2 and exon 3 sequences are optimal because they provide information on the mature HLA-DP protein, the target of T-cell recognition. The 3′UTR regulatory region variants that contribute to HLA-DP expression are in strong positive LD across the HLA-DPB1 genetic locus. The highest associations with the rs9277534 marker are located within exon 3, which are the minimal essential data required to infer high- or low-expression if the 3′UTR is not directly characterized by current HLA-DPB1–typing platforms.17 Hence, to accommodate a combined TCE and expression approach for donor selection, complete exon 2 and exon 3 sequence information is desirable.
To obtain a comprehensive catalog of HLA-DPB1 sequences, we leveraged 178 096 donors from the Be The Match Registry, whose HLA-DPB1 sequences were characterized using next-generation sequencing methods. This powerful data set provided an ultra-high sequence definition that permitted exon 2 and exon 3 variations to be phased for definitive haplotype analysis, which is required to combine TCE and expression models. A major benefit of the donor pool is its racial and ethnic diversity. In this endeavor, rich donor resources provided entirely novel population-based data on the frequency of high- and low-expression allotypes. A major finding of this study is the high frequency of high-expression allotypes in AFA and API Americans, which requires both exon 2 and exon 3 sequence data to unequivocally assign the allele. In contrast to AFA and API Americans, CAU Americans have a significantly higher frequency of low-expression allotypes. These data provide new insights into the population genetics of HLA-DPB1 and emphasize the need for complete sequence data for both exons 2 and 3 for newly discovered alleles and retrospective definitions for known alleles that lack exon 3 data. Future steps involve leveraging the 7-locus haplotype data (HLA-A∼B∼C∼DRB1∼DRB3/4/5∼DQB1∼DPB1) generated by Gragert et al (2023) to include more than 8 million US donors in this extensible analysis.36
This study did not seek to study the clinical significance of TCE, Predicted Indirectly Recognizable HLA Epitopes, and expression in the risk of GVHD, relapse, or mortality after unrelated donor transplantation. Rather, our goal was to provide users with a flexible approach to incorporate HLA-DP into prospective donor selection. The choice of which model(s) to use rests with the user. To this end, a tool should accommodate each model individually or in combination. If a sequential approach is taken, in which TCE is considered first, followed by HLA-DP expression or vice versa, an understanding of potential differences in eligible donors is desirable. We leveraged a large cohort of patients who had previously undergone an unrelated donor transplantation to define how a tool can meet needs, regardless of the order in which the models are prescribed. We show that when TCE is used as the first step to screen donors, a second step based on expression can further refine donor choices. Buhler et al demonstrated that clinical outcome after HLA-11/12 transplantation is superior when donors mismatched against a low-expression allotype in the recipient are selected; when there are multiple HLA-11/12 donors mismatched against a low-expression allotype, donors with a TCE-permissive mismatch can be considered.22 Independent studies with larger data set sizes need to be conducted to confirm these clinical validations. We envision that as the number of registry donors with upfront HLA-DPB1 typing steadily grows, more patients may have 2 or more single HLA-DPB1–mismatched donors and benefit from a 2-step selection procedure.
Our tool accommodates both TCE and expression simultaneously and, thus, can accommodate single or double HLA-DPB1 mismatches between the candidate donor and recipient. When both TCE and expression are considered simultaneously, our data suggest that the choices for preferred donors are anticipated to vary by patient race and ethnicity. Efforts to fully characterize HLA-DPB1 alleles in diverse populations across coding and noncoding regions remain important objectives for future research and will provide more complete information on both TCE groups and expression levels. This information directly implies a relationship between alleles with shared peptide-binding domain motifs (G groups). A more complete definition of the coding and regulatory variation of alleles within G groups may uncover additional variation and provide new information regarding their functional relevance.
Our overarching goal was to provide the community with an accurate and efficient means of handling complex HLA-DP TCE and its expression features in day-to-day clinical decision-making. Its design and development rely on complete sequence information to ensure that a wide range of typing data can be handled while minimizing errors in allele inference. In doing so, we uncovered new information on the shared and unique variations across racially diverse US populations. With the success in the development and refinement of transplantation regimens and procedures that increase the safety and efficacy of HLA-matched and HLA-mismatched transplantation for all patients in need, we anticipate a continued need to understand the extent of HLA variation and the implications of structure on function. For HLA-DP, a template for the addition of novel polymorphisms has been developed to facilitate clinical and research efforts.
Acknowledgments
The authors thank Pradeep Bashyal for software infrastructure, Gregory Bringman for extracting donor sequence data, and Dan Valiga for developing the program used for extracting donor sequence data.
The authors acknowledge funding from the National Cancer Institute (CA100019, CA231838, and CA218285) and the National Institute of Allergy and Infectious Diseases (AI069197) of the National Institutes of Health (E.W.P.). Bioinformatics methods development was supported in part by the Office of Naval Research grant N00014-21-1-2954 to NMDP/BTM (R.S., Y.-T.B., and M.J.M.).
R.S. is a PhD candidate at the University of Minnesota, Twin Cities. This work is submitted in partial fulfillment of the requirement for the PhD.
Authorship
Contribution: E.W.P., M.J.M., and Y.-T.B. conceived and supervised the study; R.S. developed ExPAT (UI, Python package, and analysis pipeline) and created the figures; and all authors contributed to the manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yung-Tsi Bolon, Immunobiology and Bioinformatics Research, National Marrow Donor Program/Be The Match, Center for International Blood and Marrow Transplant Research, 500 N 5th St, Minneapolis, MN 55401; e-mail: ybolon@nmdp.org.
References
Author notes
Original data from data sets 1, 2, and 3, which all contain private information, are available upon request from bioinformatics-web@nmdp.org.
The user interface and code for ExPAT and analysis are available at https://dpb1-tce-expression.nmdp.org/ and GitHub (https://github.com/nmdp-bioinformatics/dpb1-expression).
The full-text version of this article contains a data supplement.

![A multicomponent analysis of data set 3 examining TCE-expression concordance rates, distributions of TCE-expression mismatches and expression-favorable alternatives, and median HLA-11/12 (HLA-A, -B, -C, -DRB1, and -DQB1-matched 10/10 + HLA-DPB1 single mismatch) donor search list sizes. The match category shorthand includes Ex– (expression-unfavorable), Ex+ (expression-favorable), TCE+ (TCE-permissive), and TCE– (TCE-nonpermissive). (A) Cross tabulation of patients’ immunogenic TCE and expression (mis)match categories and concordance and discordance rates between both models. Concordance rates are the proportion of concordant matches (green and red) within all expression-relevant matches (total of Ex+ and Ex–). NA indicates matches that were not applicable to the expression model (patients with homozygous high [H] or low [L] expression genotypes: HH or LL). (B) Distribution of HLA-DPB1 match types (based on expression and TCE) for 2545 donor-recipient transplantations in data set 3 conducted from 2008 to 2021 for 9.2% HH, 43.1% HL, and 47.6% LL expression genotype patients, who were also stratifed based on individual SIRE categories. The expression-relevant information is outlined in gold. (C) Median list sizes of 11/12 donor search lists across SIRE categories.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/17/10.1182_bloodadvances.2022009554/2/m_blooda_adv-2022-009554-gr4.jpeg?Expires=1765897481&Signature=bAXp32UO8W3jD9WrGPY7it2MN7OJEG0RV3FnfPuYWZg6~3eGl6Wt---b5joH3sEUcMogUtGyqEruyc6WJvDrUsbDDGXpJh0nAMTBN0VInaB9DVgVrX6Q96bn-AQnHaTKKQgH70VUTs6gEOghAvTBjl9HnlYhZMmN-Yi1QQPTbs69TzBtZJpF2MPHhNfE7XX~USboejLsk7DEKay8hwNz6aSrKSWXdyJo2~Epx8mUw4js8zDwszF8Y-4SJ~Yhs4aszTxFzQMOTvYlMZ2wTWJUW6XxxGs6KplL~LX5-u~vy6k1Z3Yvkbug4uN4zLXnQ~uR4OPdFQl5UJXKxQiOdG05PA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)