Key Points
ICC and the 2022 WHO diagnostic classifications of AML present major similarities in real-world settings.
Conventional cytogenetics, usually rapidly available and low-cost, can stratify ∼56% of secondary AML cases.
Abstract
The increasing knowledge of molecular genetics of acute myeloid leukemia (AML) necessitated the update of previous diagnostic and prognostic schemes, which resulted in the development of the World Health Organization (WHO), the International Consensus Classification (ICC), and the new European LeukemiaNet (ELN) recommendations in 2022. We aimed to provide a real-world application of the new models, unravel differences and similarities, and test their implementation in clinical AML diagnosis. A total of 1001 patients diagnosed with AML were reclassified based on the new schemes. The overall diagnostic changes between the WHO 2016 and the WHO 2022 and ICC classifications were 22.8% and 23.7%, respectively, with a 13.1% difference in patients’ distribution between ICC and WHO 2022. The 2022 ICC “not otherwise specified” and WHO “defined by differentiation” AML category sizes shrank when compared with that in WHO 2016 (24.1% and 26.8% respectively, vs 38.7%), particularly because of an expansion of the myelodysplasia (MDS)-related group. Of 397 patients with a MDS-related AML according to the ICC, 55.9% were defined by the presence of a MDS-related karyotype. The overall restratification between ELN 2017 and ELN 2022 was 12.9%. The 2022 AML classifications led to a significant improvement of diagnostic schemes. In the real-world setting, conventional cytogenetics, usually rapidly available and less expensive than molecular characterization, stratified 56% of secondary AML, still maintaining a powerful diagnostic role. Considering the similarities between WHO and ICC diagnostic schemes, a tentative scheme to generate a unified model is desirable.
Introduction
Diagnosis and treatment of acute myeloid leukemia (AML) require an integrated approach that takes into account clinical and laboratory characteristics, morphologic evaluation of bone marrow and peripheral blood, flow cytometry, and cytogenetic and molecular analyses.1 The recent advances in molecular medicine have unveiled the clinical utility of genomic profiles, which necessitated the integration of this information into daily practice for both diagnostic and prognostic purposes. In 2022, 2 updated classifications and a new prognostic stratification of AML have been proposed, including the fifth edition of the World Health Organization (WHO) classification of tumors,2 The International Consensus Classification (ICC)3 of Myeloid Neoplasms and Acute Leukemias, and the European LeukemiaNet (ELN) recommendations for AML prognosis.4
Among the main innovations introduced by both diagnostic classifications, the blast threshold required for AML diagnosis is certainly one of the most notable. The limit of 20% blasts continued to be used for the majority of AML categories in the 2016 WHO classification5 (except for RUNX1::RUNX1T1-AML, CBFB::MYH11-AML, and acute promyelocytic leukemia). This threshold was discarded in the group with AML, defining recurrent genetic abnormalities based on the 2022 WHO criteria, but ICC continues to have a 10% limit.3,6 For all other AML subtypes, including myelodysplasia (MDS)-related AML (AML-MR), the 20% blast cutoff is maintained. ICC also introduced the MDS/AML subentity in cases with blasts ranging from 10% to 20%, to indicate a fading of the diagnostic boundaries between MDS and AML when judged solely based on blast proportion, thereby emphasizing the increasing importance of molecular footprints to define the various nosologic subtypes.3,6 One of the major updates of the 2022 classifications is the more precise and objective definition of secondary AML (sAML). In this context, patients’ clinical history is added as a disease attribute in the ICC and includes therapy-related AML or a prior diagnosis of MDS or MDS/myeloproliferative neoplasm. Germ line predisposition is also considered as a diagnostic qualifier, and because of its relevant frequency, it has been confirmed as a patient and disease feature requiring specific definition.3 Furthermore, the prior AML with MDS-related changes is now better defined by a specific genomic signature and referred to as AML-MR within the WHO 2022 classification. Conversely, ICC identifies multiple new subcategories, namely AML with myelodysplasia-related cytogenetic abnormalities (AML-MDSk), AML with MDS-related gene mutations (AML-MDSgene), and AML with mutated TP53 (AML-TP53).6,7
The updated 2022 ELN recommendations differ from the previous 2017 edition8 for the abrogation of the prognostic role previously assigned to FLT3-internal tandem duplication (ITD) allelic ratio (AR), based on the recognition of the favorable impact of FLT3-inhibitors on the prognosis of FLT3-ITD+ AML, now all included in the ELN intermediate-risk group.4 A further addition is the acknowledgment of the unfavorable prognostic role of the expanded list of MDS-related gene mutations, including ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2, which now define an adverse-risk ELN group.
The impact of these innovations on clinical practice is not known yet.9 Certainly, the amount of information required for the diagnostic and prognostic definitions implies a strong collaboration between clinicians and hematology laboratories. Besides, every patient should have access to standardized molecular tests, and this may not be readily feasible in all parts of the world.10 Molecular testing upon cytogenetics is particularly encouraged by the ICC hierarchical structure, whereby AML-TP53 precedes AML-MDSgene and the latter supersedes the AML-MDSk category. To investigate the clinical impact of the 2022 editions of the AML classification and prognostication systems, we reclassified a cohort of 1001 patients with AML, previously diagnosed and stratified according to WHO 2016 and ELN 2017 criteria. Our primary objective was to provide a real-world application of the new AML classifications, testing the improved disease definition compared with previous versions, and validate the 2022 ELN prognostic stratification.
Patients and methods
Patient characteristics
A total of 1001 patients with AML were included in this study after obtaining written informed consent. The main inclusion criterion was a diagnosis of AML based on the 2016 WHO classification. Patients previously classified as having MDS, with a blast count ranging from 10% to 19% were not included in this analysis. Patient data were collected through an International collaboration, which included the following: (1) Tor Vergata University, Rome, Italy (34 cases); (2) the Humanitas Cancer Center, Milan, Italy (88 cases); (3) the Cleveland Clinic Foundation, OH (466 cases); and (4) publicly available source (the BEAT-AML Master Trial; 413 cases); patients were treated between the years 2012 and 2022.11,12 With the exception of BEAT-AML Master Trial, data were obtained retrospectively via chart review. Characteristics of Cleveland Clinic and BEAT-AML cohorts have been made publicly available elsewhere (https://github.com/ardadurmaz/aml). Biological samples and chart review for this study were obtained in accordance with the Declaration of Helsinki principles and local ethics committee’s approval. At the time of initial AML diagnosis, all patients underwent bone marrow aspiration, conventional cytogenetics, and next generation sequencing (NGS) analyses, in accordance with standard guidelines.
All samples were evaluated for the mutational status of a list of commonly mutated myeloid genes, including ASXL1, BCOR, CEBPA, EZH2, FLT3, NPM1, RUNX1, SF3B1, SRSF2, STAG2, TP53, U2AF1, and ZRSR2. Mutations were detected using NGS, with a variant calling at a ≥2% threshold.11,12FLT3-ITD mutation burden was assessed either by capillary electrophoresis, as AR, or by NGS as variant allelic frequency, in order to group patients per ELN 2017.8,13 Cytogenetic analysis was performed using standard chromosome banding techniques and documented in compliance with the International System for Human Cytogenomic Nomenclature recommendations.14 Evaluation of at least 20 metaphases was required.15 Patient clinical characteristics are shown in Table 1, whereas genetic subgrouping is detailed in Figure 1.
Patient characteristics
| . | Patients with AML (n = 1001) . |
|---|---|
| Male:female ratio (M/F) | 539:462 |
| Median age, y (range) | 61 (1-93) |
| Age <65 y, n (%) | 597 (59.7) |
| Survival outcome available, n∗ (%) | 881 (92.4) |
| Treatment available,n∗(%) | 870 (91.3) |
| Conventional chemotherapy | 688 (72.2) |
| Nonintensive therapy† | 144 (15.1) |
| Palliative care | 37 (3.9) |
| FLT3-inhibitors (alone or in combination with CTX) | 107 (11.2) |
| Hematopoietic stem cell transplantation | 323 (33.9) |
| . | Patients with AML (n = 1001) . |
|---|---|
| Male:female ratio (M/F) | 539:462 |
| Median age, y (range) | 61 (1-93) |
| Age <65 y, n (%) | 597 (59.7) |
| Survival outcome available, n∗ (%) | 881 (92.4) |
| Treatment available,n∗(%) | 870 (91.3) |
| Conventional chemotherapy | 688 (72.2) |
| Nonintensive therapy† | 144 (15.1) |
| Palliative care | 37 (3.9) |
| FLT3-inhibitors (alone or in combination with CTX) | 107 (11.2) |
| Hematopoietic stem cell transplantation | 323 (33.9) |
APL, acute promyelocytic leukemia; CTX, chemotherapy; n, number.
Excluding patients with APL and those with not-available survival or treatment data.
Therapy with hypomethylating agents (azacitidine or decitabine), venetoclax, and AG-221.
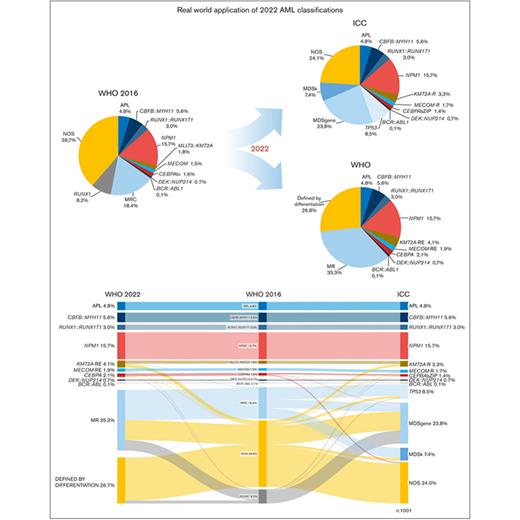
Patient’s distribution based on WHO 2016, ICC, and WHO 2022 diagnostic classifications. (A) Distribution of 1001 patients with AML based on the diverse classifications. (B) Relationship, overlaps, and differences across different AML subtypes shown using a Sankey plot. The overall proportion of genetically defined AML, based on the new classifications, is shown in brackets. APL, acute promyelocytic leukemia; MECOM-R, MECOM rearrangements; MECOM-RE, MECOM with extended rearrangements; KMT2A-R, KMT2A rearrangements; KMT2A-RE, KMT2A with extended rearrangements; MPN, myeloproliferative neoplasm; MR, myelodysplasia-related; MRC, myelodysplasia-related changes.
Patient’s distribution based on WHO 2016, ICC, and WHO 2022 diagnostic classifications. (A) Distribution of 1001 patients with AML based on the diverse classifications. (B) Relationship, overlaps, and differences across different AML subtypes shown using a Sankey plot. The overall proportion of genetically defined AML, based on the new classifications, is shown in brackets. APL, acute promyelocytic leukemia; MECOM-R, MECOM rearrangements; MECOM-RE, MECOM with extended rearrangements; KMT2A-R, KMT2A rearrangements; KMT2A-RE, KMT2A with extended rearrangements; MPN, myeloproliferative neoplasm; MR, myelodysplasia-related; MRC, myelodysplasia-related changes.
Patients were treated in accordance with local protocols, including conventional chemotherapy (688 patients), nonintensive therapy (144 patients), and best supportive care (37 patients). Tyrosine-kinase inhibitors were used, either alone or in combination with chemotherapy, in 11.2% of cases. Overall, the median follow-up time was 37.6 months (interquartile range, 14.7-73.1 months) from initial diagnosis.
Statistical analysis
Patients’ characteristics were summarized based on descriptive statistics of median and range (continuous variables) or frequencies and percentages (categorical variables).
The distribution of patients among the categories identified by the ELN classifications was studied using contingency tables, in which both absolute frequencies and percentages related to them were reported. The association between categorical variables in contingency tables was evaluated using Pearson χ2 test. The number of patients reclassified from WHO 2016 to WHO 2022 and ICC 2022 classsifications was compared using a 2-sample z test for equality of proportions with continuity correction.
Overall survival (OS) estimations with 95% confidence intervals were computed using the Kaplan-Meier method, and survival curves were compared using the log-rank test.
Two univariate Cox models were created to compare the prognostic abilities of the ELN 2017 and ELN 2022 classifications. The Akaike Information Criterion was used to compare the goodness of fit to the data of the 2 models. All tests were 2-sided, with P < .05 indicating a statistically significant difference.
Statistical analysis was performed using R software.16
Results
Distribution of AML subsets based on ICC and 2022 WHO classifications
To validate the diagnostic accuracy of the 2022 AML classifications, we reclassified a cohort of 1001 patients based on the 2022 WHO and ICC models.2,3Table 2 shows the main differences among WHO 2016, WHO 2022, and ICC classifications, whereas Figure 1 shows the distribution of the diagnostic subcategories based on the different classifications. In general, the overall shift of diagnostic categories was 22.8% and 23.7% between WHO 2016 vs WHO 2022 classifications and ICC, respectively, whereas this was 13.1% between ICC and WHO 2022 classification. The size of categories defined as “not otherwise specified” (NOS) by ICC and “defined by differentiation” (DD) according to WHO 2022 significantly shrank when compared with that in WHO 2016 classification (24.1% and 26.8%, respectively, vs 38.7%; P < .0001), particularly because of an expansion of MDS-related categories. According to the ICC, older patients were more frequently diagnosed with AML-TP53 or AML-MDSgene (supplemental Figure 1).
AML classifications
| AML . | WHO 2016 . | ICC . | WHO 2022 . |
|---|---|---|---|
| With defining genetic abnormalities∗ | PML::RARA (APL)† | RARA-R (APL) | PML::RARA (APL) |
| RUNX1::RUNX1T1† | RUNX1::RUNX1T1 | RUNX1::RUNX1T1 | |
| CBFB::MYH11† | CBFB::MYH11 | CBFB::MYH11 | |
| MLLT3::KMT2A | KMT2A-R | KMT2A-RE | |
| MECOM | MECOM-R | MECOM-RE | |
| BCR::ABL1 (provisional entity) | BCR::ABL1‡ | BCR::ABL1‡ | |
| NPM1 | NPM1 | NPM1 | |
| CEBPAbi | CEBPAbZIP | CEBPA‡ | |
| DEK::NUP214 | DEK::NUP214 other rare recurring translocations# | DEK::NUP214 | |
| RBM15::MKL1 | RBM15::MRTFA | ||
| Previous history of MDS, or MDS/MPN§ | MRC | MR | |
| disease defining | diagnostic qualifiers | disease defining | |
| Myelodysplasia-related with defining cytogenetic abnormalities§ | MDSk | ||
| complex karyotype | complex karyotype | complex karyotype | |
| -7/del(7q), del(5q)/t(5q), i(17q)/t(17p), -13/del(13q), del(11q), del(12p)/t(12p), idic(X)(q13) | del(5q)/t(5q)/add(5q),-7/del(7q), +8, del(12p)/t(12p)/add(12p), i(17q), -17/add(17p) or del(17p), del(20q), idic(X)(q13) | del5q or 5q, loss-7 or del7q or 7q loss, del11q, del12p or 12p loss, -13 or del13q, del17p or 17p loss or i17q, idic(X)(q13) | |
| balanced abnormalities | |||
| Dysplasia§ | >50% of cells of at least 2 lineages | ||
| With defining mutations§ | RUNX1 (provisional entity) | MDSgene | |
| ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2 | ASXL1, BCOR, EZH2, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2 | ||
| TP53-mutǁ | TP53 | ||
| Without defining genetic abnormalities¶ | NOS | NOS | DD (defined by differentiation) |
| AML . | WHO 2016 . | ICC . | WHO 2022 . |
|---|---|---|---|
| With defining genetic abnormalities∗ | PML::RARA (APL)† | RARA-R (APL) | PML::RARA (APL) |
| RUNX1::RUNX1T1† | RUNX1::RUNX1T1 | RUNX1::RUNX1T1 | |
| CBFB::MYH11† | CBFB::MYH11 | CBFB::MYH11 | |
| MLLT3::KMT2A | KMT2A-R | KMT2A-RE | |
| MECOM | MECOM-R | MECOM-RE | |
| BCR::ABL1 (provisional entity) | BCR::ABL1‡ | BCR::ABL1‡ | |
| NPM1 | NPM1 | NPM1 | |
| CEBPAbi | CEBPAbZIP | CEBPA‡ | |
| DEK::NUP214 | DEK::NUP214 other rare recurring translocations# | DEK::NUP214 | |
| RBM15::MKL1 | RBM15::MRTFA | ||
| Previous history of MDS, or MDS/MPN§ | MRC | MR | |
| disease defining | diagnostic qualifiers | disease defining | |
| Myelodysplasia-related with defining cytogenetic abnormalities§ | MDSk | ||
| complex karyotype | complex karyotype | complex karyotype | |
| -7/del(7q), del(5q)/t(5q), i(17q)/t(17p), -13/del(13q), del(11q), del(12p)/t(12p), idic(X)(q13) | del(5q)/t(5q)/add(5q),-7/del(7q), +8, del(12p)/t(12p)/add(12p), i(17q), -17/add(17p) or del(17p), del(20q), idic(X)(q13) | del5q or 5q, loss-7 or del7q or 7q loss, del11q, del12p or 12p loss, -13 or del13q, del17p or 17p loss or i17q, idic(X)(q13) | |
| balanced abnormalities | |||
| Dysplasia§ | >50% of cells of at least 2 lineages | ||
| With defining mutations§ | RUNX1 (provisional entity) | MDSgene | |
| ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2 | ASXL1, BCOR, EZH2, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2 | ||
| TP53-mutǁ | TP53 | ||
| Without defining genetic abnormalities¶ | NOS | NOS | DD (defined by differentiation) |
Although for WHO 2016 a blast count ≥ 20% is required for AML diagnosis, ICC requires ≥ 10% blasts; for WHO 2022, no minimal blast counts is required.
KMT2A-R, KMT2A rearrangements; KMT2A-RE, KMT2A with extended rearrangements; MECOM-R, MECOM rearrangements; MECOM-RE, MECOM with extended rearrangements; MPN, myeloproliferative neoplasm; MR, myelodysplasia-related; MRC, myelodysplasia-related changes; mut, mutation.
Major differences among classifications are highlighted in bold.
For WHO 2016 a blast count ≥ 20% is required; for ICC a blast count ≥ 10% is required;for WHO 2022 no blast count is required. Exceptions are indicated by the other symbols.
No minimal blast count required; exceptions to the WHO 2016 classification.
Blast count ≥ 20% is still required; exceptions to the ICC and WHO 2022 classifications.
A blast count ≥ 20% is still required.
A blast count ≥ 20% is required.
A blast count ≥ 20% is still required.
PRDM16::RPN1, NPM1::MLF1, KAT6A::CREBBP, RBM15::MRTFA, NUP98, and other partners, ETV6::MNX1, PICALM::MLLT10, FUS::ERG, RUNX1::CBFA2T3, and CBFA2T3::GLIS2.
Setting the WHO 2016 classification as a backbone to compare the shift of nosologic categories (Figure 1B), the number of KMT2A-rearranged and MECOM rearrangements with AML was lower in ICC than WHO 2022 classification (5.0% vs 6.0%), but it was still higher than that in the WHO 2016 classification (3.3%; supplemental Figure 2) because of a precise definition of KMT2A and MECOM atypical rearrangements in ICC. The CEBPA-mutated AML category, which included 1.6% cases with biallelic-CEBPA (CEBPAbi) mutations in WHO 2016 classification, included 1.4% CEBPAbZIP mutated cases based on ICC. This category was broad in the WHO 20222 classification because of the inclusion of both CEBPAbi and CEBPAbZIP mutations (2.1%). RUNX1-mutated AML, indicated by the WHO 2016 classification as a provisional entity, was eliminated in the new classifications. As a result, 82 RUNX1-mutated AML cases were reclassified based on the WHO 2022 classification as follows: 74.4% as AML-MR, 22.0% as AML-DD, 2.4% as AML-CEBPA, and 1.2% as AML-MECOM with extended rearrangements. In the ICC, 92.7% of cases were classified as AML-MDSgene, 3.7% as AML-TP53, 2.4% as AML-CEBPAbZIP, and 1.2% as MECOM rearrangements with AML.
Molecular profile vs cytogenetics for the diagnosis of sAML
According to the different criteria proposed by the 2 schemes (Table 2), we reclassified the subgroups of sAML (Figure 1). The 184 cases previously defined as AML with MDS-related changes increased to 353 AML-MR according to WHO 2022 classification (35.3% of our cohort), whereas ICC criteria led to the identification of 312 AML-MR cases (23.8% AML-MDSgene and 7.4% AML-MDSk) and 85 TP53-mutated AML cases (8.5%).
Only 4 patients with AML-MR based on the WHO 2022 classification were defined having NOS according to the ICC, because of the presence of 11q- and 13q-cytogenetic abnormalities (each in 2 patients), and these were not included in the ICC AML-MDSk category (supplemental Figure 3). Contrastingly, 90.6% of ICC AML-TP53 cases were reclassified to the WHO 2022 AML-MR group because of the concomitant presence of a complex karyotype in 81.8% cases, a MDS-related gene mutation in 7.8% cases, a MDS-related cytogenetic abnormality, or both a cytogenetic and a molecular alteration in 10.4% cases.
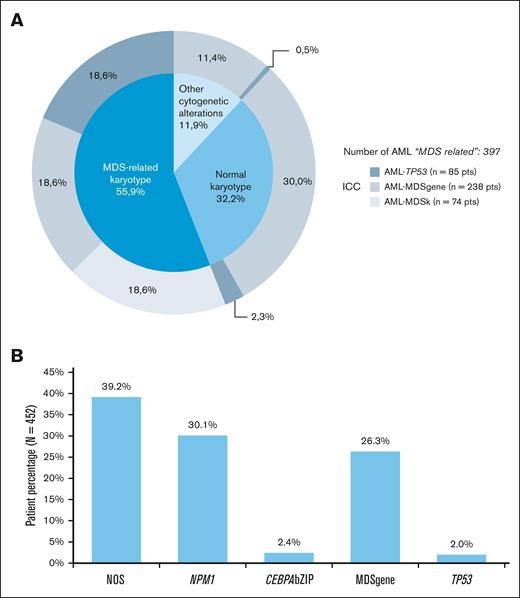
We then explored whether a reversed ICC hierarchical approach, with cytogenetics as the first and NGS as a second step, could also be used to assign patients to the different subgroups. Out of 397 patients with AML-MR per the ICC, 55.9% cases (222 patients) were defined by the presence of a MDS-related karyotype, including those grouped in the AML-TP53, AML-MDSgene, and AML-MDSk categories based on ICC (Figure 2A).
Distribution of patients with AML based on their cytogenetic and molecular profile. (A) Distribution of secondary AML (n = 397 AML with MDS-related categories), based on the presence of MDS-related cytogenetic abnormalities. Each category was further stratified according to the hierarchical ICC classification system, shown in the outer circle (AML with TP53 mutation, AML with myelodysplasia-related gene mutations, and AML with myelodysplasia-related cytogenetic abnormalities: gray shades). (B) Frequency of ICC diagnostic subcategories in patients with AML with NK (n = 452).
Distribution of patients with AML based on their cytogenetic and molecular profile. (A) Distribution of secondary AML (n = 397 AML with MDS-related categories), based on the presence of MDS-related cytogenetic abnormalities. Each category was further stratified according to the hierarchical ICC classification system, shown in the outer circle (AML with TP53 mutation, AML with myelodysplasia-related gene mutations, and AML with myelodysplasia-related cytogenetic abnormalities: gray shades). (B) Frequency of ICC diagnostic subcategories in patients with AML with NK (n = 452).
When considering AML with normal karyotype (NK; n = 452), 34.5% patients presented ≥1 ICC MDS–related gene mutation, but these mutations had a diagnostic relevance in only 26.3% of cases according to the ICC hierarchical algorithm (Figure 2B). Furthermore, 34.5% patients presented with ≥1 additional AML-diagnostic gene mutation (NPM1, 30.1%; CEBPAbZIP, 2.4%; and TP53, 2.0%). Therefore, NGS refined the diagnosis in 60.8% and 58.0% of patients with NK AML based on ICC and WHO 2022 classification, respectively.
Risk stratification based on ELN
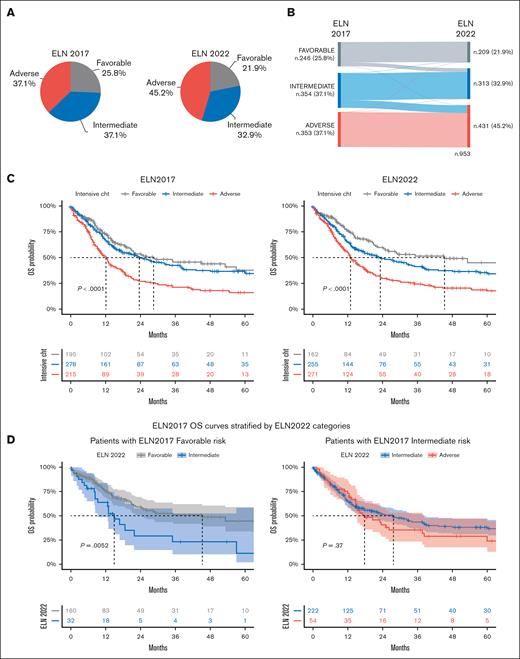
According to the ELN 2017 prognostic stratification, patients with AML (n = 953 patients, excluding 48 patients with acute promyelocytic leukemia) were divided into the following 3 subgroups: 25.8% as favorable, 37.1% as intermediate, and 37.1% as adverse. By applying the ELN 2022 criteria, in which FLT3-ITD mutations invariably identify intermediate risk and 9 somatic mutations define unfavorable risk (Figure 3A), 21.9% of patients were placed in the favorable-risk, 32.9% in the intermediate-risk, and 45.2% in the adverse-risk groups (P < .05 for the favorable- and intermediate-risk groups and P < .0005 for the adverse-risk group). Overall, the changes in restratification between ELN 2017 and ELN 2022 resulted in a reclassification of 12.9% of the patients.
AML stratification based on the ELN 2017 and 2022. (A) Stratification of patients with AML (n = 953, excluding patients with acute promyelocytic leukemia). (B) Relationship between risk groups, as shown using a Sankey plot, comparing the 2 ELN models. (C) Kaplan-Meier curves show survival estimates of patients with AML treated with conventional chemotherapy according to ELN 2017 and 2022 (n = 688 patients with available survival data). (D) OS of patients with favorable- (n = 192) and intermediate-risk (n = 276) AML, based on ELN 2017, treated with conventional chemotherapy and restratified by ELN 2022. Among ELN 2017 favorable-risk cases, 3 were restratified into the adverse-risk group based on ELN 2022 and were, therefore, not included. Furthermore, 2 patients belonging to the ELN 2017 intermediate-risk category were not included, because they were restratified into the ELN 2022 favorable-risk group. (E) OS of patients with NK AML treated with conventional chemotherapy (n = 74), classified in the ICC AML-MDSgene category, and based on the presence of 1 or ≥2 MDS-gene mutations. Data of patients alive at last follow-up were censored. Numbers of those at risk are indicated below the curves and are color-coded. P values shown are the result of the log-rank test.
AML stratification based on the ELN 2017 and 2022. (A) Stratification of patients with AML (n = 953, excluding patients with acute promyelocytic leukemia). (B) Relationship between risk groups, as shown using a Sankey plot, comparing the 2 ELN models. (C) Kaplan-Meier curves show survival estimates of patients with AML treated with conventional chemotherapy according to ELN 2017 and 2022 (n = 688 patients with available survival data). (D) OS of patients with favorable- (n = 192) and intermediate-risk (n = 276) AML, based on ELN 2017, treated with conventional chemotherapy and restratified by ELN 2022. Among ELN 2017 favorable-risk cases, 3 were restratified into the adverse-risk group based on ELN 2022 and were, therefore, not included. Furthermore, 2 patients belonging to the ELN 2017 intermediate-risk category were not included, because they were restratified into the ELN 2022 favorable-risk group. (E) OS of patients with NK AML treated with conventional chemotherapy (n = 74), classified in the ICC AML-MDSgene category, and based on the presence of 1 or ≥2 MDS-gene mutations. Data of patients alive at last follow-up were censored. Numbers of those at risk are indicated below the curves and are color-coded. P values shown are the result of the log-rank test.
All patients with AML classified as being at adverse risk based on ELN 2017 had no change in the risk class in the 2022 edition (Figure 3B), with the exception of 2 patients (1 with high FLT3-ITD AR and the other with RUNX1 and a monoallelic-CEBPAbZIP mutations). Of the ELN 2017 intermediate-risk group, 77.4% remained in this category, whereas 0.8% and 21.8% moved into the favorable- and adverse-risk groups, respectively (Figure 3B). Of 77 patients with AML with an ELN 2017 intermediate karyotype, 76 were reclassified to the adverse-risk group in the 2022 revision because of the presence of ≥1 MDS-related gene mutation, whereas 1 presented with an atypical MECOM rearrangement.
Looking at the ELN 2017 favorable-risk group, 83.3% of AML cases were confirmed as having favorable risk based on ELN 2022, whereas 15.5% and 1.2% were upstaged into the ELN 2022 intermediate– and adverse–risk groups, respectively. Considering the AML recategorized as an intermediate-risk group according to ELN 2022, 34 of 38 patients were diagnosed as having AML-NPM1/FLT3-ITD–low, whereas the remaining 4 patients presented with CEBPAbi mutations not involving the bZIP domain.
Regarding outcomes, both ELN editions confirmed their stratification capability, without significant differences in their prognostic power, as defined in the Akaike Information Criterion (supplemental Table 2). A total of 688 patients treated using conventional chemotherapy were evaluated, and the 24-month OS was 53.6% and 59.7% in the favorable-risk, 49.7% and 49.0% in the intermediate-risk, and 27.0% and 30.0% in the adverse-risk AML categories for ELN 2017 and ELN 2022, respectively (Figure 3C). After restratifying per ELN 2022, for the 192 patients classified as being at favorable risk per ELN 2017 and treated with conventional chemotherapy, the difference in the OS between favorable- and intermediate-risk groups was statistically significant (P < .01), whereas there was no survival difference between patients reclassified as being at adverse risk, deriving from the ELN 2017 intermediate risk (Figure 3D). Contrastingly, both ELN 2017 and ELN 2022 did not stratify patients treated with nonintensive therapies (supplemental Figure 5).
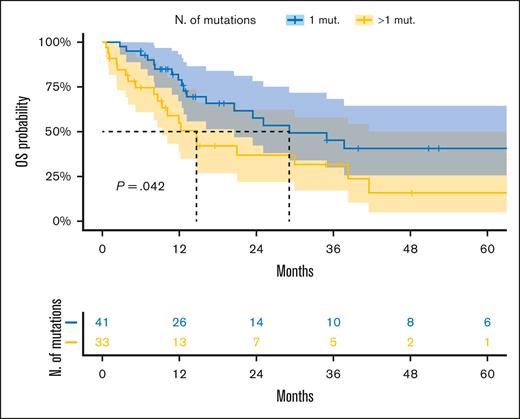
We also focused on the heterogeneous group of patients with NK and adverse mutations per the ELN classification. Specifically, patients treated with intensive regimens with multiple MDS-related gene mutations had significantly worse outcomes than those harboring only 1 MDS-related gene lesion (Figure 3E).
Discussion
In 2022, 2 new AML classifications were proposed as a response to the new molecular advances in the field. Our extensive real-word reclassification of 1001 patients confirmed the crucial role of the newly added molecular alterations for an accurate AML diagnosis. Indeed, NGS helped precisely recategorize AML cases previously categorized in the NOS “basket category,” mainly by expanding the MDS-related group (82.7% in the WHO 2022 MR category and 78.7% in the MDSgene/TP53 ICC categories).17,18 Notwithstanding this, it must be recognized that although a prominent role of NGS analysis is given by the ICC hierarchic structure, this may not be readily available at all centers.
Herein, we discuss some prototypic examples. Looking at the rarer AML subtypes, KMT2A and MECOM atypical rearrangements and CEBPAbZIP single-mutation, expanded the AML-defining disease categories in both the 2022 classifications. Given the importance of these abnormalities, their correct identification becomes essential. One of the most important differences between the 2022 classifications is the introduction of AML-TP53 disease category in the ICC because of the negative prognostic role of biallelic TP53 mutations (or ≥10% variant allelic frequency) regardless of blast counts.7,19 We found that ∼90% of AML with TP53 mutations were included in the AML-MR based on the WHO 2022 classification because of the coexistence of cytogenetic alterations, especially complex karyotype (81.8% of cases), and/or accompanying somatic MDS-related gene mutations (supplemental Figure 3).7 In this context, 9.4% of AML-TP53, according to the ICC, would not emerge based on WHO 2022 rules. Harmonization between the 2 new classifications toward the diagnostic role of TP53 in AML is crucial, especially once a TP53-targeted therapy will hopefully be available.20
Another difference is represented by the case of RUNX1-mutated AML, which is an AML-defining mutation in the ICC, but not in the 2022 WHO classification, and is prognostically unfavorable in ELN 2022. However, of 76 patients with RUNX1-mutated AML in our cohort, classified as having AML-MDSgene in the ICC, 77.6% were included in the WHO 2022 AML-MR category because of the presence of ≥1 additional MDS-related gene mutation.19 Given the association of RUNX1 mutations with sAML and the evolution of bone marrow failure syndromes, examining for these mutations in the diagnostic process of AML may need to be reconsidered.21,22
Although the criteria to define AML-MR based on previous history of MDS or MDS/myeloproliferative neoplasm were maintained in the WHO 2022 classification, the panel of MDS-related genes, together with MDS cytogenetic alterations, covers most of sAML, regardless of their previous history.11 Indeed, the genetic signature of these entities appears similar to that of high-risk MDS and generally of AML progressing from antecedent hematologic disorders, as compared with de novo AML.11,23,24 Additionally, the diagnostic classifiers become of prognostic significance as the MDS-gene signature identifies cases with adverse ELN risk, as shown by previous studies focusing on AML ontogeny.11,17,25,26 Although it has been shown that the time from AML diagnosis to treatment start does not significantly affect the outcome in clinically stable patients, the currently proposed diagnostic algorithm posits some methodological and socioeconomic challenges.27 Indeed, it must be considered that NGS analysis may not be widely available and often needs long turnaround times and prohibitive costs at some centers. The main issue concerns MDS-related AML in which the hierarchical approach, according to ICC, requires the availability of the mutation status before or simultaneously with karyotype information, which is far from reality, even in many experienced, high-resource centers. In NK AML, NGS indeed helped refine the diagnosis in 60.8% and 58.0% of the patients, based on the ICC and WHO 2022 classification, respectively, whereas more than half of the patients with AML-MR (56%) could be characterized via conventional cytogenetics and stratified as being at adverse risk. Therefore, conventional cytogenetics demonstrated to be a useful tool, still able to discriminate a large number of patients in a fast, relatively low-cost, and easier fashion. Contrastingly, NGS proves to be essential to correctly classify ∼44% of the patients with AML-MR. In this context, concerted efforts are now needed to make NGS more cost effective and globally available, especially in cases in which it changes the treatment approach. In this line, the appeal to dedicated referral centers highly specialized in NGS diagnostics may help harmonize the process of AML diagnosis.
The ELN 2022 risk stratification, regardless of the relevant adjustments, did not improve the prognostic capability compared with the earlier version in our patients. This may be related to the conceptually important but relatively unsubstantial changes between the 2 schemes, the heterogeneity of treatments adopted in our real-world cohort, and the use of strategies partly belonging to the pre-FLT3 inhibitors era.28
One of the most important updates in ELN 2022 is the reconsideration of FLT3-ITD AR, whose role has been abolished.29 Therefore, the presence of mutated FLT3-ITD without any other adverse genetic abnormalities defines the intermediate risk, regardless of FLT3-ITD AR and the copresence of NPM1 mutations. In our cohort, the majority of patients considered as being at ELN 2017 favorable risk who were restratified as being at ELN 2022 intermediate risk consisted of NPM1-mutant/FLT3-ITD–low. The accuracy of ELN 2022 in identifying favorable-risk AML was confirmed with the use of our real-word data, with the clinical consequence of avoiding the overtreatment of patients. However, capillary electrophoresis continues to be recommended as the standardized diagnostic tool for FLT3 mutations,30 particularly for its quick turnaround and in light of the indication for treatment with FLT3-inhibitors such as midostaurin as well as for its ability to detect longer size FLT3-ITDs, which are underestimated in NGS.31
The newly defined adverse-risk group, enriched with participants with NK and ≥1 adverse mutation, deserves a special mention. Our data confirm the reports from a previous study that patients with a single MDS-related gene mutation have a better OS than those with >1 alteration.32 Thus, one could speculate whether having a single MDS-related gene and a NK is sufficient to decide upon treatment intensification, as suggested in a recent ELN 2022 validation study.33 Future efforts focusing on the relation between the new prognostication system and various treatments (especially hematopoietic stem cell transplantation) are warranted.
This study presented some caveats. In particular, the retrospective and multicenter nature of our patient cohort determined that some results might have been affected by differences in health care systems and practice as well as the broad time range in patient enrollment. Furthermore, all AML cases were defined by the presence of ≥20% blasts, excluding the new ICC MDS/AML category from this evaluation.
In conclusion, the use of molecular data is now crucial to precisely diagnose and risk-stratify patients with AML. More than a dozen genes have been incorporated within current diagnostic and prognostic schemes, and genome scanning approaches have been proposed to capture the complexity of AML biology. However, the high costs, expertise level in molecular biology, and issues of standardization still represent hurdles to provide equitable care for patients with AML worldwide. Given the substantial similarities, the conflicts between the newly competing diagnostic systems may be resolved, reaching an agreement on a unified model, which must also take into account the available resources worldwide.
Acknowledgments
This work was supported by AIRC 5 × 1000 call “Metastatic disease: the key unmet need in oncology” to MYNERVA project, #21267 (Myeloid Neoplasms Research Venture AIRC. A detailed description of the MYNERVA project is available at http://www.progettoagimm.it). This work was also supported by MUR-PNRR M4C2I1.3 PE6 project PE00000019 Heal Italia, PRIN grant 2017WXR7ZT, Ministero della Salute, Rome, Italy (Finalizzata 2018, NET-2018-12365935; personalized medicine program on myeloid neoplasms: characterization of the patient’s genome for clinical decision making and systematic collection of real-world data to improve quality of health care) (M.T.V.), and the AIRC Foundation (Associazione Italiana per la Ricerca contro il Cancro, Milan Italy; project #22053) (M.G.D.P.). C.G. was supported by a grant from the Edward P. Evans Foundation.
Authorship
Contribution: E.A., A.S., and M.T.V. designed the study, interpreted the data, and wrote the manuscript; B.B. took part in manuscript writing; C.G. collected data, edited the manuscript, helped in data interpretation, and gave helpful intellectual insights during the study; A.P. and M.C. performed statistical analysis; T.O., E.F., S.T., M.D., M.R.P., and A.D. helped in data analysis and participated in data interpretation; H.A., V.V., J.P.M., M.G.D.P., and A.V. participated in samples and data collection; M.T.V. took responsibility for the integrity and the accuracy of the data presented; and all authors reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carmelo Gurnari, Department of Biomedicine and Prevention, Tor Vergata University, Rome, Italy; e-mail: carmelogurnari31@gmail.com.
References
Author notes
∗E.A. and A.S. contributed equally to this study.
Data used for this study can be found in GitHub for open-source access at https://github.com/ardadurmaz/aml and a publicly available source (BEAT-AML Master trial).
Additional information is available on request from the corresponding author, Carmelo Gurnari (carmelogurnari31@gmail.com).
The full-text version of this article contains a data supplement.