TO THE EDITOR:
Next-generation immunotherapies for multiple myeloma (MM) that target B-cell maturation antigen (BCMA, also known as TNFRSF17, BCM, CD269, and TNFRSF13A), including monoclonal antibodies, bispecific T-cell engagers, and chimeric antigen receptor (CAR) T cells, have shown remarkable benefits for patients in clinical studies, but relapse still occurs.1-4 Resistance to immunotherapy can occur via tumor extrinsic or intrinsic mechanisms, with extrinsic mechanisms usually related to changes in immunosuppressive cells, immunosuppressive cytokines, coinhibitory receptors, and costimulatory receptors in the tumor microenvironment.5-11 The intrinsic mechanisms include antigen loss due to mutations,12,13 deletions,14,15 and splicing pattern changes.16,17 Of these 2 mechanisms, tumor intrinsic mechanisms tend to be rarer. However, 2 recent studies found that biallelic loss of BCMA causes resistance to anti-BCMA therapy.12,14 Although BCMA is a critical target for immunotherapy, the incidence of heterozygous and homozygous deletions of BCMA has previously been reported only in smaller cohorts of newly diagnosed patients and patients with relapsed myeloma, and it is unknown whether they correlate with other genomic alterations.14,18-20 Here, we evaluated 2883 patients with MM at diagnosis and relapse to understand the frequency of BCMA-targeting somatic events and the characteristics of patients with MM with a BCMA deletion.
We analyzed patient data from 4 existing cohorts of newly diagnosed patients with MM to evaluate monoallelic BCMA loss events in MM. Single nucleotide polymorphism (SNP) arrays and whole genome sequencing (WGS) samples were collected from patients with MM after they signed an informed consent form, and the study was accepted by the Toulouse ethics committee. The clinical and genomic data were deidentified in accordance with the Declaration of Helsinki and genotyped according to their cohort protocols.
The SNP array analysis was performed as following. Genomic DNA purified from primary MM samples was interrogated with Affymetrix CytoScan HD, SNP 6.0, or 500K Arrays (Affymetrix, Santa Clara, CA) according to the manufacturer’s instructions. Smoothed copy number (CN) estimates for each probe set were exported from processed CEL files. The CN estimates were calculated as the average CN of all probe sets within each cytoband region.20
For WGS analysis, we performed 150 bp paired-end sequencing on HiSeqX10 genome analyzers. Homozygous/heterozygous deletions, copy-neutral loss-of-heterozygosity, allele-specific gains/amplification, and the ploidy and purity of each sample were analyzed using fraction- and allele-specific CN estimates from tumor sequencing.21
We downloaded the CoMMpass data set from The Multiple Myeloma Research Foundation data portal. Segmented CN variant and point mutation calls were used.
We identified the frequency of deletions using segmented CN alteration (CNA) calls and genomic identification of significant targets in cancer (GISTIC).22 Segmented files generated with fraction- and allele-specific copy number estimates from tumor sequencing or downloaded from the MMRF data portal were converted into text files and analyzed using GISTIC. For each gene, the amplification and deletion frequencies were extracted from the GISTIC output files.
Single cells were encapsulated using a 10X chromium controller (10X Genomics) according to the chromium single-cell DNA user guide (10X Genomics). The reads were aligned to the human reference genome hg38. Cells were called, and CNs were estimated using the Cell Ranger-DNA “cnv” command with default options.
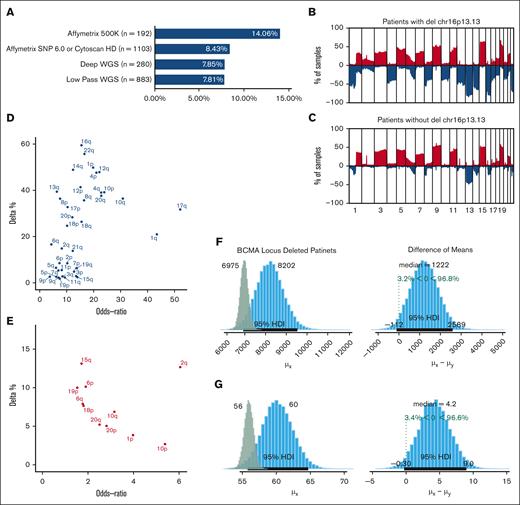
We evaluated the frequency of deletions involving the BCMA locus (16p13.13) in 2458 newly diagnosed patients with MM from multiple studies using data from SNP arrays and shallow and deep WGS. We observed monoallelic del16p in 8.58% of the newly diagnosed patients (n = 2458; Figure 1A). The observed frequency varied for each study, ranging from 7.81% to 14.6%, and both SNP array- and sequencing-based methods showed similar results (supplemental Figure 1).
Monosomy 16p events in Multiple Myeloma. (A) BCMA deletion frequency (x-axis) according to each study. The number of patients enrolled in each data set is shown next to the platform used. (B-C) Top graph shows the frequency of CNAs in patients with del16p (B) and patients without del16p (C) across all chromosomes (blue = loss and red = gain). The y-axis shows the frequencies, and the x-axis shows 1 mb of the genomic locus from the beginning of chromosome 1 to chromosome 22. (D) The difference in the frequency of deletions (y-axis) and OR (x-axis) of various chromosomal arms between patients with del16p13.13 and those without. (E) Difference in the frequency of gains (y-axis) and OR (x-axis) of various chromosomal arms between patients with del16p13.13 and those without. (F-G) Posterior distributions of mutational load between newly diagnosed patients with del16p (blue bars) and those without (green bars) in the WGS dataset (F) and MMRF (G) cohorts. The median numbers are shown at the top of each panel, and 95% credible intervals are shown with black lines at the bottom of the distribution. Difference of means between the 2 groups and the posterior probabilities are shown in the right panels.
Monosomy 16p events in Multiple Myeloma. (A) BCMA deletion frequency (x-axis) according to each study. The number of patients enrolled in each data set is shown next to the platform used. (B-C) Top graph shows the frequency of CNAs in patients with del16p (B) and patients without del16p (C) across all chromosomes (blue = loss and red = gain). The y-axis shows the frequencies, and the x-axis shows 1 mb of the genomic locus from the beginning of chromosome 1 to chromosome 22. (D) The difference in the frequency of deletions (y-axis) and OR (x-axis) of various chromosomal arms between patients with del16p13.13 and those without. (E) Difference in the frequency of gains (y-axis) and OR (x-axis) of various chromosomal arms between patients with del16p13.13 and those without. (F-G) Posterior distributions of mutational load between newly diagnosed patients with del16p (blue bars) and those without (green bars) in the WGS dataset (F) and MMRF (G) cohorts. The median numbers are shown at the top of each panel, and 95% credible intervals are shown with black lines at the bottom of the distribution. Difference of means between the 2 groups and the posterior probabilities are shown in the right panels.
Next, we evaluated genome-wide CNAs. We found that the frequency of BCMA loss did not differ between patients with hyperdiploid and nonhyperdiploid MM and that the overall CN loss was significantly higher in patients with BCMA loss (Figure 1B-C). The CN loss in these patients affected most of the chromosomes at varying frequencies, whereas chromosomal gains were limited in scope (Figure 1D-E). High-risk deletion events, such as del1p and del17p, were frequently observed in patients with BCMA loss compared with those in other patients (odds ratio [OR], 19.37; 95% confidence interval [CI], 13.13-25.80); false discovery rate = 1.57e–65 vs OR, 8.8 [CI, 6.39-12.15]; false discovery rate = 5.57E–39, respectively; supplemental Tables 1-2). However, high-risk CN gains were not different between patients with and without 16p loss. Similar to CN events, patients with BCMA loss had a higher mutational load (8202 with 95% high-density interval (HDI) 6921-9535) compared with those without (6975 with 95% HDI 6626-7343). In brief, patients with BCMA loss had a 96.8% chance of having more mutations (difference of the means = 1222 [95% CI, –112 to 2589]; Figure 1F). Similar differences were observed in the MMRF cohort using deep whole-exome sequencing (Figure 1G). However, BCMA loss should not be considered the cause of these increases; it may be a consequence of other high-risk events, such as increased genomic instability.
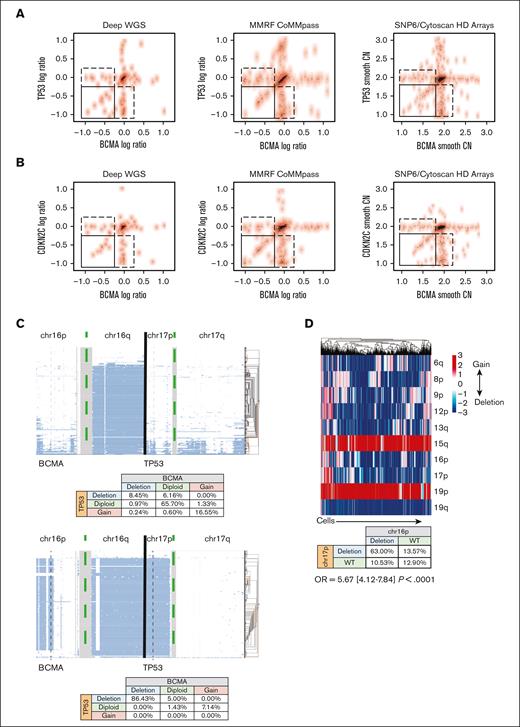
To understand the risk profile of patients with BCMA loss, we focused on the observation that BCMA loss frequently co-occurs with other deletions. In fact, when BCMA and TP53 losses were present in the same patient, they had similar log ratios (sequencing) or smoothed CNs (SNP array; Figure 2A). Similarly, when using CDKN2C as a proxy for chromosome 1p loss, we observed that when both BCMA and CKDN2C loss were present, they had similar CN values (Figure 2B). These data suggested the co-occurrence of these events in the same cells when both events were observed in the samples. However, it should also be noted that not every patient with del17 or del1p carried del16p or vice versa. Therefore, we used single-cell DNA sequencing for 2 patients with BCMA loss, 1 with subclonal (8.45%) and the other with clonal loss, and found that almost all cells with a BCMA deletion had a TP53 deletion also (Figure 2C), whereas not all cells with TP53 loss had BCMA loss. This suggested that TP53 loss occured first in this single case, followed by BCMA loss. We further investigated whether this held true in a case of biallelic BCMA loss (after anti-BCMA, targeted CAR T-cell therapy), using single-cell RNA sequencing data to impute CNAs. We found that BCMA loss still tended to co-occur with TP53 deletions (OR, 5.67; 95% CI, 4.12-7.84; P < .0001; Figure 2D).
Correlation between monosomy 16p and high risk events. (A-B) The CN of BCMA and TP53 within each patient across various genomic platforms, assessed using bulk genomic data. Log ratio of sequencing coverage is shown on the left and middle panels for WGS data sets. Smooth copy number estimated from the SNP array is used on the right panels. Top panels show BCMA and TP53 locus (A) for each patient (a dot in the scatter plot) in 3 data sets, and bottom panels (B) show BCMA and CDKN2C loci. Spearman rank correlation ρ for samples in the bottom left corner box (samples in which both genes are deleted) is 0.65 (P value = 6.27e–06) for the WGS data sets and 0.53 (P value = 2.6e–05) for SNP array in the top panels and 0.67 (P = 8.67 e−08) and 0.65 (P = 7.8e–07) in the bottom panels, respectively. (C) CN estimates of the single cells (each row in panels) for 2 patients (right and left panels). Clusters or single cells are shown with histograms on the right of each panel. Genomic regions from the beginning of the chromosome 16p arm to the end of the chromosome 17 q arm are shown on the x-axis. The coverage and data quality are shown in the bottom panels. The co-occurrences of CN events are shown in the bottom tables for each patient. Vertical gray lines are used to mark BCMA and TP53 locations. (D) (Top) Graphs showing the co-occurrence of deletions (blue) and gains (red) of CNAs at the single-cell level in a relapsed patient. Each column is a cell. (Bottom) Table showing the co-occurrence of deletions and duplications of BCMA and TP53.
Correlation between monosomy 16p and high risk events. (A-B) The CN of BCMA and TP53 within each patient across various genomic platforms, assessed using bulk genomic data. Log ratio of sequencing coverage is shown on the left and middle panels for WGS data sets. Smooth copy number estimated from the SNP array is used on the right panels. Top panels show BCMA and TP53 locus (A) for each patient (a dot in the scatter plot) in 3 data sets, and bottom panels (B) show BCMA and CDKN2C loci. Spearman rank correlation ρ for samples in the bottom left corner box (samples in which both genes are deleted) is 0.65 (P value = 6.27e–06) for the WGS data sets and 0.53 (P value = 2.6e–05) for SNP array in the top panels and 0.67 (P = 8.67 e−08) and 0.65 (P = 7.8e–07) in the bottom panels, respectively. (C) CN estimates of the single cells (each row in panels) for 2 patients (right and left panels). Clusters or single cells are shown with histograms on the right of each panel. Genomic regions from the beginning of the chromosome 16p arm to the end of the chromosome 17 q arm are shown on the x-axis. The coverage and data quality are shown in the bottom panels. The co-occurrences of CN events are shown in the bottom tables for each patient. Vertical gray lines are used to mark BCMA and TP53 locations. (D) (Top) Graphs showing the co-occurrence of deletions (blue) and gains (red) of CNAs at the single-cell level in a relapsed patient. Each column is a cell. (Bottom) Table showing the co-occurrence of deletions and duplications of BCMA and TP53.
Collectively, our data show that even without treatment pressure, BCMA monoallelic deletions are frequent events. Moreover, patients with these events have an increased deletion frequency of other chromosomes, making these cells more vulnerable to the biallelic loss of BCMA and other genes. In fact, these events tended to occur in the same cells. Events and treatments may select for such cells, thus increasing the presence of high-risk clones and impacting subsequent treatment options. Future studies are needed to investigate whether antibody treatments select for these cells and dual antigen-targeting therapies provide benefits to such cases. One observation from our study was that most of these events were subclonal, such that the bulk assessments of messenger RNA or protein expression normally taken in the clinic may miss this loss (supplemental Figure 2). However, the presented data are limited to monoallelic losses and do not suggest that these patients are going to develop biallelic losses or that these monoallelic deletions are the cause of other high-risk events. In fact, monoallelic 16p deletions are potential consequences of other factors, such as genomic instability. Therefore, combining strategies at the bulk and single-cell level are necessary to not only better understand the biology but also a particular patient’s illness.
Acknowledgments: This study was supported by National Institutes of Health grants P01 CA155258 and P50 CA100707, Paula and Rodger Riney Foundation, and VA Healthcare System grant 5I01BX001584.
Contribution: M.K.S. and N.M. designed the research; M.K.S., A.A.S., R.L., and P.S. analyzed the data; and all authors contributed to data collection and wrote the manuscript.
Conflict-of-interest disclosure: K.A. has received consulting fees from Janssen, Pfizer, AstraZeneca, and Daewoong and served on the board of directors of and had stock options in C4 Therapeutics, OncoPep, Window, Dynamic Cell Therapies, and Starton. N.M. is consultant for Bristol Myers Squibb, Janssen, OncoPep, Amgen, Karyopharm, Legend, AbbVie, Takeda, and GlaxoSmithKline and has stock options in OncoPep. The remaining authors declare no competing financial interests.
Correspondence: Mehmet Kemal Samur, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: mehmet_samur@dfci.harvard.edu; and Nikhil Munshi, Department of Data Science, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: nikhil_munshi@dfci.harvard.edu.
References
Author notes
MMRF data are available via the MMRF research portal at https://research.themmrf.org.
Single nucleotide polymorphism array data are available in the Gene Expression Omnibus database (accession number GSE12896).
The sequencing data set is available in the European Genome-phenome Archive (accession number EGAS00001006731).
Additional data are available on request from the corresponding author, Nikhil Munshi (nikhil_munshi@dfci.harvard.edu).
The full-text version of this article contains a data supplement.