Key Points
Identification of human bone marrow mesenchymal cells with osteopontin (SPP1) overexpression in patients with MDS.
SPP1 expression in comparable mesenchymal stromal cell populations plays protective roles in disease progression in an MDS mouse model.
Abstract
Myelodysplastic syndromes (MDSs) are a heterogenous group of diseases affecting the hematopoietic stem cell that are curable only by stem cell transplantation. Both hematopoietic cell intrinsic changes and extrinsic signals from the bone marrow (BM) niche seem to ultimately lead to MDS. Animal models of MDS indicate that alterations in specific mesenchymal progenitor subsets in the BM microenvironment can induce or select for abnormal hematopoietic cells. Here, we identify a subset of human BM mesenchymal cells marked by the expression of CD271, CD146, and CD106. This subset of human mesenchymal cells is comparable with mouse mesenchymal cells that, when perturbed, result in an MDS-like syndrome. Its transcriptional analysis identified Osteopontin (SPP1) as the most overexpressed gene. Selective depletion of Spp1 in the microenvironment of the mouse MDS model, Vav-driven Nup98-HoxD13, resulted in an accelerated progression as demonstrated by increased chimerism, higher mutant myeloid cell burden, and a more pronounced anemia when compared with that in wild-type microenvironment controls. These data indicate that molecular perturbations can occur in specific BM mesenchymal subsets of patients with MDS. However, the niche adaptations to dysplastic clones include Spp1 overexpression that can constrain disease fitness and potentially progression. Therefore, niche changes with malignant disease can also serve to protect the host.
Introduction
Myelodysplastic syndromes (MDSs) are age-associated clonal diseases of the hematopoietic stem cells (HSCs) characterized by bone marrow (BM) failure, cytopenias, and leukemic transformation in one-third of the affected individuals.1,2 MDS is a multistage process of hematopoietic transformation, often, starting with clonal hematopoiesis and ending with overt leukemia.3-5 However, it remains unclear what governs disease progression because many of the associated mutations do not confer a clonal competitive advantage in animal models.6,7 Acquisition of secondary mutations8 can clearly drive transition, but animal models also suggest that an altered hematopoietic microenvironment may contribute to or select for abnormal hematopoietic cells. This may be particularly relevant in the context of aging and inflammation.9-11 For example, specific genetic alterations in stromal cells, including Dicer1, Sbds, or Ptpn11, are sufficient to induce hematopoietic dysplasia and leukemia. Remarkably, mutations specifically within the immature mesenchymal stromal populations (but not within the mature osteoblasts) resulted in the development of MDS that transformed into acute myeloid leukemia (AML) in the case of Dicer1, Sbds, and β-Catenin12,13 and juvenile myelomonocytic leukemia in the case of Ptpn11.14 These data point to the important interplay that exists between specific subsets of mesenchymal cells in the niche and the hematopoietic cells that they support throughout adult tissue maintenance.
Does this same interplay exist in human patients with MDS? The BM mesenchymal abnormalities that occur outside the aforementioned experimental settings of the niche-induced oncogenesis remain undefined. It is known that the mesenchymal cells of the BM are organized in a stem cell–based hierarchy without mature cell division to replenish cells upon injury or under homeostatic conditions.15 The turnover time of the mesenchymal bone cells is measured in weeks or months.15 Therefore, somatic mutations occurring in a BM stem or progenitor cell can be expected to propagate the defect to its progeny, creating a field of mutant cells. These cells can abnormally produce cytokines that can affect hematopoietic cells such as the alarmins, S100A8/9, which Zambetti et al16 showed in a mouse model of Schwachman-Diamond syndrome. They further showed that the S1008/9 interaction with TLR4 can induce genotoxic stress in hematopoietic stem and progenitor cells (HSPCs) and that increased production of S100A8/9 by stromal cells associates with worse clinical outcome in patients with MDS.16 These data suggest the potential for cooperativity between alterations in the hematopoietic cells and stroma.
The effects of abnormal HSPCs on neighboring stromal cells has also been defined. This includes evidence for an altered differentiation status of osteolineage cells17 and remodeling of the niche18 by AML cells and by conversion of mesenchymal cells to fibrogenic myofibroblasts in myeloproliferative disease models.19
These murine model systems were engineered to have genetic alterations in different and specific mesenchymal cells including those identified by the expression of osterix (Osx),20 nestin (Nes),21Prx1,22,23 leptin receptor (Lepr),24Mx1,15 and collagen I (Col1).12 These data indicate that the type of mesenchymal cell that was genetically altered differentially affects the hematopoietic phenotype.12,13
To validate these murine models in humans, we sought to define mesenchymal niche abnormalities in patients with MDS. Firstly, we defined the human mesenchymal subsets that demonstrated characteristics similar to that of the mouse mesenchymal cells shown to drive a MDS phenotype, specifically, the Osx+ mesenchymal progenitor cells. Gene expression analysis was performed on the Osx+ equivalent cells in 16 patients with MDS and 11 age-matched control samples without MDS (patients undergoing hip replacement surgery). This was paired with gene expression analysis of CD34+CD38− HSCs for a subset of the patients. To assess the effects of those gene expression perturbations identified in the mesenchymal stromal subsets of patients, we created a chimeric animal model in which the Vav-driven NUP98-HOXD13 (NHD13) fusion transgene competes with wild-type (WT) BM in the context of a microenvironment in which a specific gene of interest has been deleted. The NHD13 fusion gene, originally identified in a patient with therapy-related AML,25 leads to a fatal progressive MDS in transgenic animals.26 The predictable and progressive nature of disease in this model provides an opportunity to test how stromal perturbations affect MDS disease kinetics.
Methods
Collection of mesenchymal stromal cells from BM filter bags and hip replacement BM samples
Filter bags
BM filter bags from healthy adult, eligible BM donors were rinsed with phosphate buffered saline (PBS) to collect leftover BM. The filters were cut and put in a 50 mL conical with 10 mL of collagenase type 1 (0.25%) (Stem cell technologies, Ref# 07902) and incubated at 37°C in a shaking water bath (120 rpm) for 1 hour. Cells from the BM bags and the filters were then pooled, washed with PBS, and counted using acridine orange.
Hip replacement BM
The BM was diluted in PBS, strained using 100 μm cell strainer, and counted using acridine orange. For both, BM filter bags and hip replacement samples, 5 μL/mL of the RosetteSep pre-enrichment cocktail (Stem cell technologies, Ref#15896C) was added to a cell suspension of 40 × 106 cells per mL and incubated at room temperature (RT) for 10 minutes. Cells were then layered on ficoll and centrifuged at 400g for 25 minutes with the breaks turned off. Mononuclear cells were then washed, counted, and frozen in fetal bovine serum (FBS) supplemented with 10% dimethyl sulfoxide.
BM samples of patient with MDS
Frozen BM mononuclear cells were received from multiple clinical centers. On the day of the sorting, frozen cells were thawed in 37°C water bath, added to a tube containing 250 μL of 2000 U/mL DNAse (Sigma, Ref# D4513), and diluted with PBS up to 10 mL. Cells were centrifuged at 300g for 10 minutes at RT.
Staining for flow cytometry analysis and cell sorting
Cells were stained for 30 minutes at a concentration of 200 × 106 cells per mL in PBS supplemented with 2% FBS, 100 U/mL DNAse, and the following antibodies: CD271 (ME20.4), CD146 (P1H12), CD31 (WM-59), CD235ab (HIR2), CD45 (HI30), CD106 (STA), CD34 (581), and 7-aminoactinomycin D (7AAD) for viability.
For gene expression analysis, between 100 and 1000 cells per population were sorted using BD FACS ARIA II directly into RLT plus lysis buffer for subsequent RNA extraction, and analysis was performed using FlowJo software.
Fibroblast colony-forming unit and in vitro differentiation
For the single-cell colony-forming assay, single cells were sorted into flat bottom 96-well plate prefilled with water in the peripheral wells and with Mesencult-XF media (Stemcell Technologies) in the middle wells. Plates were then incubated under 5% CO2 at 37°C from 12 to 15 days, after which the wells were scored for the presence of colonies and confluency using a score from 1 to 3, in which 1 is the least confluent and 3 is the most confluent.
For in vitro differentiation, colonies that scored 3 were detached using MesenCult-ACF enzymatic dissociation and MesenCult-ACF enzyme inhibition solution (Stemcell Technologies), expanded in 6-well plates and differentiated into the osteogenic, adipogenic, or chondrogenic lineages using the MesenCult osteogenic or adipogenic differentiation kit (Stemcell Technologies) and human mesenchymal stem cell (MSC) chondrogenic differentiation medium BulletKit and transforming growth factor β 3 (Lonza) according to the manufacturer’s instructions. Osteogenic and adipogenic differentiation was assessed using alizarin red and oil red O staining, respectively. Chondrogenic differentiation was assessed using Sybrgreen quantitative polymerase chain reaction for Col2A1 and ACAN with the following primers:
Col2A1-F, TGGACGCCATGAAGGTTTTCT
Col2A1-R, TGGGAGCCAGATTGTCATCTC
ACAN-F, GTGCCTATCAGGACAAGGTCT
ACAN-R, GATGCCTTTCACCACGACTTC
Animal models
All animal experiments were approved by the institutional animal care and use committee at Massachusetts General Hospital. WT CD45.2 (C57BL/6J), CD45.1 (B6.SJL-Ptprca Pepcb/ BoyJ), NHD1326 (C57BL6-Tg (Vav1-NUP98/HOXD13)G2Apla/J), and Spp1 knock-out (Spp1-KO)27 (B6.129S6(Cg)-SPP1tm1Blh/J) mice were purchased from the The Jackson laboratory. The CD45-STEM (C57BL/6-CD45.1(STEM)),28 Ocn-GFPTopaz,29 Osx-GFP:Cre30 were previously described.
For the total BM transplant experiments, mice received 2× (6 Gy) doses from a cesium-137 irradiator within a 4-hour period. On the next day, 1 × 106 of the total BM mix of NHD13:CD45.2-STEM at a ratio of 3:1 were injected retro-orbitally. Male donor mice were aged from 14 to 16 weeks. Male recipient Spp1-KO or WT CD45.2 mice were used at 8 weeks of age. Peripheral blood analysis was performed at indicated time points via retro-orbital bleeding. Transplants using the antibody drug conjugate (ADC) conditioning strategy were performed as previously described.31 Briefly, biotinylated cKit Ab was purchased from BioLegend (Clone 2B8). The ADC was prepared by combining the biotinylated antibody with streptavidin-SAP (2.8 Saporin molecules per streptavidin from Advanced targeting systems) in a 1:1 molar ratio. Mice were dosed with the ckit-SAP antibody conjugate at 3 mg/kg and transplanted with a 1:1 mix of NHD13:WT cells (10 × 106 cells). Complete blood counts were obtained using the element-HT5 peripheral blood analyzer from Heska. For chimerism analysis, 100 μL peripheral blood was added to 1 mL Ack lysis buffer (Quality Biological) and incubated at RT for 7 minutes. Blood was then centrifuged at 2000g for 5 minutes and stained with the following for flowcytometry analysis: CD45.2 (104), CD45.1 (A20), CD11b-AF700 (M1/70), CD3e (145-2C11), CD45R (RA3-6B2), GR1 (RB6-8C5), and 7AAD for viability. At least 20 × 103 events were collected in the scatter gate per sample using the BD LSRII analyzer. Analysis was performed using the FlowJo software.
Collection of Osx-GFP and Ocn-GFP labeled cells for RNA sequencing
Tibias, femurs, hips, humeri, and spines were collected from Osx-GFP:Cre and Ocn-GFPTopaz hemizygous mice. BM was flushed and discarded, and bones were cut into small pieces that were digested using 0.25% collagenase type I (Stemcell Technologies) for 45 minutes in a shaking water bath at 37°C. Cells were then pelleted and washed with PBS and 2% FBS, after which they were stained with antibodies against CD-45 (30F-11), CD-31 (MEC13.3) and Ter119 (Ter119). Cells (CD45–CD31–Ter119–GFP+) were sorted using BD FACS ARIA II for subsequent RNA extraction and sequencing.
Bleeding and peripheral blood analysis
Mice were serially subjected to retro-orbital bleeding under isoflurane anesthesia. White blood cell, hemoglobin, red blood cell (RBC), mean corpuscular volume, platelet, and hematocrit levels were quantified using the blood analyzer (Heska, Element HT5). For flow cytometry for chimerism, analysis was performed on RBC–lysed blood, stained with antibodies against CD45.1 (A20), CD45.2 (104), CD11b (M1/70), GR1 (RB6-8C5), CD45R (RA3-6B2), and CD3 (145-2C11). Samples were then analyzed using BD LSRII, and data were analyzed using FLowJo.
RNA extraction
RNA extraction was performed using the AllPrep DNA/RNA Micro Kit (Qiagen, Ref# 80284) according to the manufacturer’s instructions.
RNA-seq
RNA sequencing (RNA-seq) libraries were constructed using the Clontech SMARTER kit (Takara) and sequenced on the Illumina HiSeq2500 instrument, resulting in ∼30 million reads per sample on average.
RNA-seq analysis
Transcriptome mapping was performed using the STAR aligner32 and the hg19 assembly of human reference genome. Read counts for individual transcripts from ENSEMBL annotation were obtained using HTSeq.33 Differential expression analysis was performed using the EdgeR package34 after normalizing read counts and including only genes with counts per million >1 for at least 1 sample. Differentially expressed genes (DEGs) were defined based on the criteria of more than a twofold change in normalized expression value and false discovery rate (FDR) < 0.05.
Results
The identification of a human mesenchymal progenitor subset that overlaps with murine mesenchymal progenitors whose alteration can promote MDS
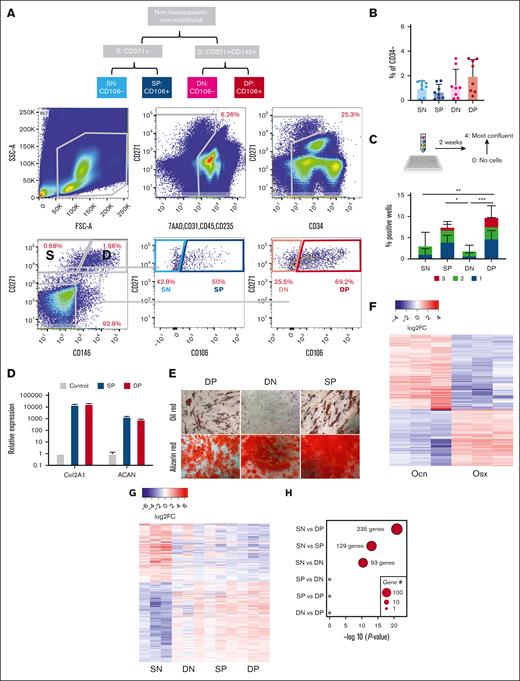
Discarded BM collection bags and filters were used as a source of human MSCs. Cells from the BM and digested bone spicules removed from the filter were stained for fluorescence-activated cell sorting (FACS). Cell surface receptors were used to mark functionally distinct BM mesenchymal subsets that have been shown to support BM hematopoiesis, namely nerve growth factor receptor (CD271), vascular cell adhesion molecule (CD106),35,36 and melanoma cell adhesion molecule (CD146).37,38 Hematopoietic, endothelial, and dead cells were removed from analysis (staining for 7AAD, CD45, CD235ab, CD31, and CD34). Within the remaining cells, the combination of CD271, CD146, and CD106 resolved 4 distinct populations: CD271+CD146−CD106− (SN), CD271+CD146−CD106+ (SP), CD271+CD146+CD106− (DN), and CD271+CD146+CD106+ (DP) (Figure 1A; supplemental Figure 1A) which were all comparably abundant within the live, nonhematopoietic nonendothelial compartment (Figure 1B).
Overlap between a human mesenchymal progenitor subset with murine mesenchymal progenitors whose alteration can promote MDS. (A) Gating strategy for the prospective isolation of mesenchymal subsets in human BM filters. Cells are a mix of BM and digested bone spicules. (B) Frequency of the distinct mesenchymal subsets within the live nonhematopoietic, nonendothelial cells. (C) Frequency of wells testing positive for cell growth of sorted single cells. Wells are scored on a scale from 1 (least confluent) to 3 (most confluent). Data represent 6 independent samples and are presented as mean ± standard deviation (SD). Indicated significance was calculated using 2-way analysis of variance; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (D) Relative expression of Col2A1 and ACAN in SP and DP cells differentiated into the chondrogenic lineage. (E) Oil and alizarin red staining for adipogenic and osteogenic differentiation of human BM or spicules mesenchymal subsets. (F) Heatmap of DEGs between Ocn-labeled osteoblasts and Osx-labeled osteoprogenitors; more than twofold change; FDR < 0.05. (G) Heatmap of DEGs between human mesenchymal subsets (3 independent samples for each); more than twofold change; FDR < 0.05. (H) Overlap of DEGs between different human mesenchymal subsets with DEGs between Ocn- and Osx-labeled mouse mesenchymal cells. x-axis shows the statistical significance of the overlap shown as P value on a log scale. Dot size represents the number of overlapping genes.
Overlap between a human mesenchymal progenitor subset with murine mesenchymal progenitors whose alteration can promote MDS. (A) Gating strategy for the prospective isolation of mesenchymal subsets in human BM filters. Cells are a mix of BM and digested bone spicules. (B) Frequency of the distinct mesenchymal subsets within the live nonhematopoietic, nonendothelial cells. (C) Frequency of wells testing positive for cell growth of sorted single cells. Wells are scored on a scale from 1 (least confluent) to 3 (most confluent). Data represent 6 independent samples and are presented as mean ± standard deviation (SD). Indicated significance was calculated using 2-way analysis of variance; ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (D) Relative expression of Col2A1 and ACAN in SP and DP cells differentiated into the chondrogenic lineage. (E) Oil and alizarin red staining for adipogenic and osteogenic differentiation of human BM or spicules mesenchymal subsets. (F) Heatmap of DEGs between Ocn-labeled osteoblasts and Osx-labeled osteoprogenitors; more than twofold change; FDR < 0.05. (G) Heatmap of DEGs between human mesenchymal subsets (3 independent samples for each); more than twofold change; FDR < 0.05. (H) Overlap of DEGs between different human mesenchymal subsets with DEGs between Ocn- and Osx-labeled mouse mesenchymal cells. x-axis shows the statistical significance of the overlap shown as P value on a log scale. Dot size represents the number of overlapping genes.
In a single-cell colony-forming assay, colonies were scored for confluency ranging from 1, which is the least confluent, to 3, which is the most confluent. CD106+ populations (SP and DP) produced significantly more colonies than CD106− counterparts and were the only populations with confluent colonies (scoring 3), indicating a faster rate of proliferation (Figure 1C). Similarly, CD106+ single-cell–derived colonies were the only colonies that efficiently differentiated toward osteogenic, adipogenic, and chondrogenic lineages in vitro (Figure 1D-E). Chondrogenic lineage differentiation was only assessed in CD106+ populations because of the limited cell numbers in the other populations. Altogether, this is indicative of higher progenitor activity in CD106+ labeled populations.
To identify the human mesenchymal subsets that demonstrated progenitor characteristics similar to those previously defined as the population that induced myelodysplasia in mice, we compared gene expression among mouse osteoprogenitor cells and mature osteoblasts sorted from genetic reporter models expressing green fluorescent protein (GFP) under the transcriptional control of osterix (Osx-GFP:Cre)30 and osteocalcin (Ocn-GFPTopaz),29 promoters specific to osteoprogenitor cells and mature osteoblasts, respectively (supplemental Figure 1B). In total, 1145 genes were differentially expressed more than twofold) at an FDR < 0.05 (Figure 1F; supplemental Table 1). In parallel, 4 human mesenchymal subsets were analyzed similarly (DP, DN, SP, and SN). Gene expression confirmed the mesenchymal identity of the human subsets in comparison with the human CD45+ cells, as demonstrated by the expression of mesenchymal specific genes (supplemental Figure 1C), as part of a total of 2223 DEGs among the 4 populations (Figure 1G; supplemental Table 2). Differential gene expression analysis was performed, comparing the mouse subsets (Ocn-GFP vs Osx-GFP) and the human populations (SN vs DP, SN vs SP, SN vs DN, SP vs DN, SP vs DP, and DN vs DP) to investigate the overlap in DEGs (Figure 1H). Comparing SN vs DP yielded the highest level of differentially expressed orthologous gene overlap (235 genes), followed by SN vs SP (129 genes) and SN vs DN (93 genes; Figure 1H), indicating that mouse Ocn-Osx cell transcriptional difference is most reminiscent of human SN-DP difference and suggesting a higher similarity of the SN and DP/SP human populations to murine Ocn– and Osx–labeled cells, respectively. As a control for the used methodology, we ran the same analysis on the mCD51 vs mCD45 and hCD51 vs hCD45, given that CD45 labels hematopoietic cells and that CD51 labels a wide range of mesenchymal cells in mouse and human BM. This comparison yielded a highly significant overlap (P < 1e−100) in 2419 of 3802 DEGs, validating our adopted strategy (supplemental Figure 1D).
Distinct gene expression alterations characterize specific BM mesenchymal subsets in patients with MDS
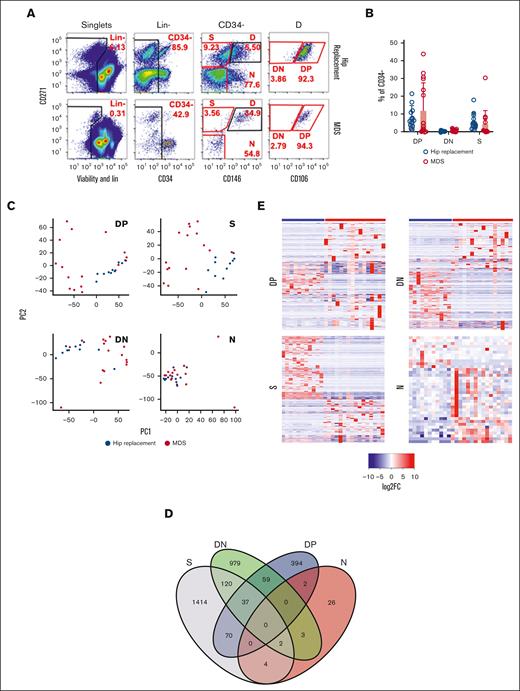
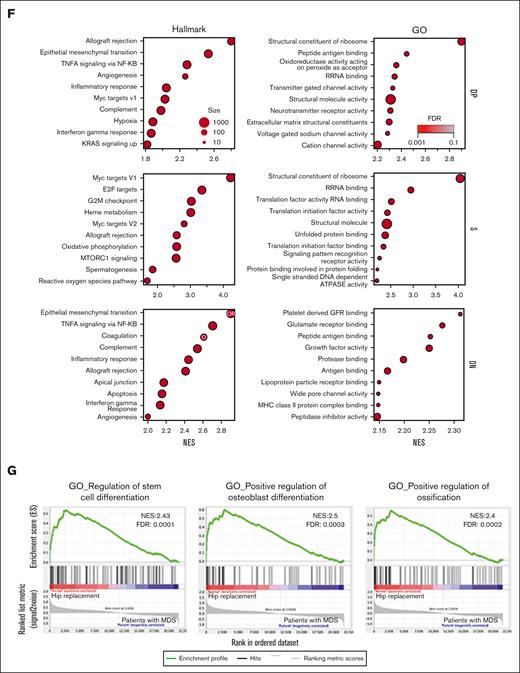
We proceeded to analyze the mesenchymal compartment of 16 BM samples of patient with MDS and 11 age- and gender-matched hip replacement samples (supplemental Table 3). The patient samples were cryopreserved aspirate samples without BM spicules and, therefore, contained fewer stromal cells. Preliminary analysis demonstrated a lower frequency of the DN population and a nonexistent SN population as compared with the BM filter bags analyzed in Figure 1, indicating that these populations are enriched in cells obtained from digested bone spicules (data not shown). Based on this, we sorted 4 populations for bulk RNA-seq: S, which combines SP and SN (because of the low frequency of the SN in the BM samples with no spicules), DN, and DP, in addition to CD146−CD271− cells (N) (Figure 2A). No significant differences in the frequency of S, DN, and DP populations within the nonhematopoietic nonendothelial compartment were detected (Figure 2B). Transcriptome-wide principal component analysis demonstrated a high degree of overlap between MDS and hip replacement samples within the N population. In the DP, DN, and S populations, hip replacement samples had a tight distribution, whereas MDS samples were separate and more scattered, indicating a distinct transcriptional profile with high degree of interpatient variability, which was most pronounced in DP and S populations (Figure 2C). Differential gene expression analysis revealed minimal overlap between the N and DP, S, and DN populations (Figure 2D), with 256 genes for S, 100 for DP, 166 for DN, and 11 for N with more than twofold change and FDR < 0.05 (Figure 2E; supplemental Table 4). Gene set enrichment analysis (GSEA)39,40 using hallmark and gene ontology gene sets demonstrated the enrichment of an inflammatory and epithelial-to-mesenchymal transition signature in the DP and DN populations of patients with MDS, whereas the S population demonstrated an enrichment in Myc targets, MTORC signaling, reactive oxygen species pathway, and ribosome and protein translation pathways (Figure 2F). Interestingly, the S population demonstrated a downregulation of functions related to stem cell and osteoblastic differentiation (Figure 2G).
Distinct gene expression signature for mesenchymal subsets in patients with MDS. (A) Gating strategy for sorting distinct mesenchymal subsets from BM of patients undergoing hip replacement therapy or patients with MDS. Red gates indicate populations sorted for sequencing. (B) Frequency of mesenchymal subsets demonstrates no significant differences between patients undergoing hip replacement and patients with MDS. (C) Transcriptome-wide principal component analysis (PCA) of distinct mesenchymal subsets demonstrating the separation between patients undergoing hip replacement surgery and patients with MDS for populations DP, S, DN, and N. (D) Venn diagram demonstrating the overlap in DEGs between patients undergoing hip replacement surgery and those with MDS in the different mesenchymal subsets. (E) Heatmaps of DEGs (more than twofold change; FDR < 0.05) in distinct mesenchymal subsets between patients undergoing hip replacement surgery and patients with MDS. (F) Functional gene categories enriched in DP, DN, and S subsets of patients with MDS. (G) GSEA demonstrating decreased osteogenic differentiation in the S subset of patients with MDS. GO, gene ontology
Distinct gene expression signature for mesenchymal subsets in patients with MDS. (A) Gating strategy for sorting distinct mesenchymal subsets from BM of patients undergoing hip replacement therapy or patients with MDS. Red gates indicate populations sorted for sequencing. (B) Frequency of mesenchymal subsets demonstrates no significant differences between patients undergoing hip replacement and patients with MDS. (C) Transcriptome-wide principal component analysis (PCA) of distinct mesenchymal subsets demonstrating the separation between patients undergoing hip replacement surgery and patients with MDS for populations DP, S, DN, and N. (D) Venn diagram demonstrating the overlap in DEGs between patients undergoing hip replacement surgery and those with MDS in the different mesenchymal subsets. (E) Heatmaps of DEGs (more than twofold change; FDR < 0.05) in distinct mesenchymal subsets between patients undergoing hip replacement surgery and patients with MDS. (F) Functional gene categories enriched in DP, DN, and S subsets of patients with MDS. (G) GSEA demonstrating decreased osteogenic differentiation in the S subset of patients with MDS. GO, gene ontology
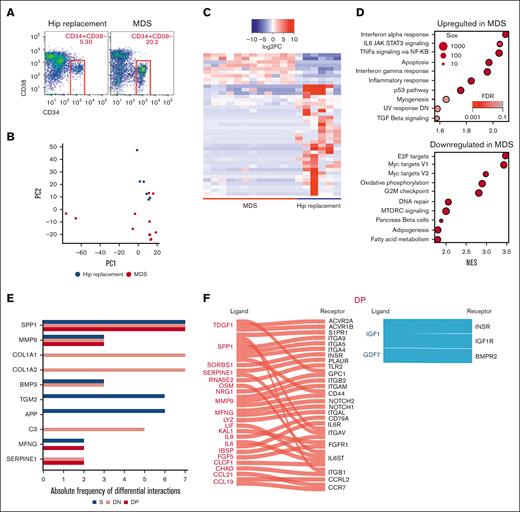
Identification of potential molecular interactions between mesenchymal subsets and MDS-propagating cells
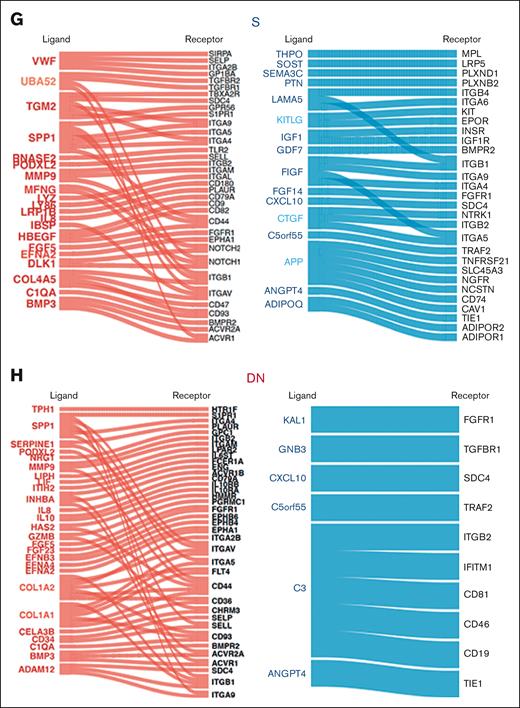
To dissect the effect of mesenchymal stromal transcriptional perturbations on MDS-propagating hematopoietic cells,2 we profiled the transcriptomes of 12 BM CD34+CD38− cells of patients with MDS and 6 of those of patients with hip replacement (Figure 3A) using the same platform and the same patient samples as those in the prior mesenchymal stromal populations analysis, when possible (9 of 12 patients with MDS and 6 of 6 patients with hip replacement; supplemental Table 3). Transcriptome-wide principal component analysis demonstrated a separation of MDS from the hip replacement samples, except for 2 samples of patients with MDS that clustered with the controls (Figure 3B). Differential gene expression analysis demonstrated 42 genes with more than twofold change and FDR < 0.05 (Figure 3C; supplemental Table 4). GSEA using hallmark gene sets demonstrated the upregulation of apoptosis, tumor necrosis factor α signaling via NF-κB, interleukin-6 JAK-STAT signaling, and inflammatory response gene sets in patients with MDS and the downregulation of MTORC1 signaling, E2F targets, DNA repair, and G2M checkpoint gene sets (Figure 3D). To identify potential mechanisms of molecular interactions between the analyzed MSC populations and the CD34+CD38− cells, we used a published database of annotated ligands and their cognate receptors.41 As potential mediators of these cell-cell interactions, we focused on the ligand-receptor pairs in which the ligand was differentially expressed in mesenchymal cells of patients with MDS compared with that in controls (more than twofold expression change, with FDR < 0.05), and the receptor was expressed in CD34+CD38− hematopoietic cells (reads per kilobase of transcript per million reads mapped [RPKM] > 1). COL1A genes in addition to SPP1 showed the highest number of interactions (Figure 3E) with SPP1 appearing in all 3 populations (Figure 3F-H; supplemental Table 5).
Identification of signaling channels between mesenchymal subsets and MDS-propagating cells. (A) Sorting strategy of CD34+CD38− cells. (B) Transcriptome-wide PCA demonstrating the separation between CD34+CD38− cells in patients undergoing hip replacement surgery and patients with MDS. (C) Heatmap of DEGs (more than twofold change; FDR < 0.05) between CD34+CD38− of patients with MDS and those undergoing hip replacement surgery. (D) Functional gene categories enriched in CD34+CD38− from patients with MDS compared with patients undergoing hip replacement surgery, according to GSEA. (E) Bar plot of ligands with highest frequency of inferred ligand-receptor interactions in the different mesenchymal subsets in which the ligand was differentially expressed in mesenchymal cells of patients with MDS compared with that in cells of the control (more than twofold expression change, with FDR < 0.05), and the receptor was expressed in CD34+CD38− hematopoietic cells. (F-H) Ligand-receptor interactions in the different mesenchymal subsets in which the ligand is differentially expressed in the indicated mesenchymal subset (red: upregulated; blue: downregulated), and the receptor is expressed (RPKM > 1) in CD34+CD38− cells.
Identification of signaling channels between mesenchymal subsets and MDS-propagating cells. (A) Sorting strategy of CD34+CD38− cells. (B) Transcriptome-wide PCA demonstrating the separation between CD34+CD38− cells in patients undergoing hip replacement surgery and patients with MDS. (C) Heatmap of DEGs (more than twofold change; FDR < 0.05) between CD34+CD38− of patients with MDS and those undergoing hip replacement surgery. (D) Functional gene categories enriched in CD34+CD38− from patients with MDS compared with patients undergoing hip replacement surgery, according to GSEA. (E) Bar plot of ligands with highest frequency of inferred ligand-receptor interactions in the different mesenchymal subsets in which the ligand was differentially expressed in mesenchymal cells of patients with MDS compared with that in cells of the control (more than twofold expression change, with FDR < 0.05), and the receptor was expressed in CD34+CD38− hematopoietic cells. (F-H) Ligand-receptor interactions in the different mesenchymal subsets in which the ligand is differentially expressed in the indicated mesenchymal subset (red: upregulated; blue: downregulated), and the receptor is expressed (RPKM > 1) in CD34+CD38− cells.
Loss of Spp1 in the nonhematopoietic microenvironment enhances the fitness of MDS cells
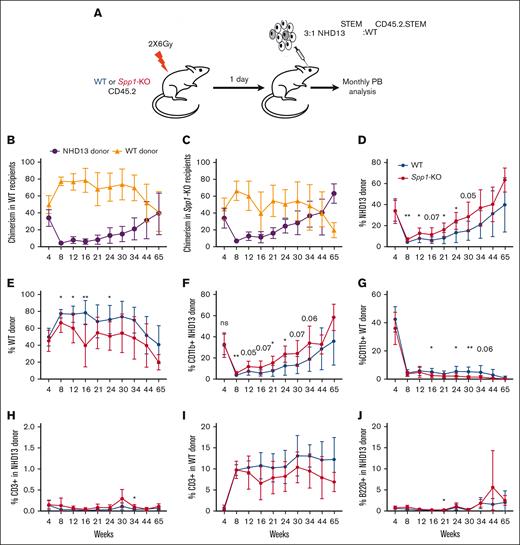
Secreted phosphoprotein 1 (SPP1), also known as Osteopontin, is a phosphorylated glycoprotein initially identified in the bone42 and later found to be produced by a plethora of cell types, including osteoclasts, epithelial cells, endothelial cells, nerve cells, vascular smooth muscle cells, and lymphoid and myeloid immune cells.43 In the hematopoietic microenvironment, we and others have shown that Spp1 acts as a negative regulator of the HSC pool size.44,45 Although it may constrain HSC numbers, it appears to preserve their function.46Spp1 expression in the BM decreases with age, leading to a decline in HSC engraftment potential. Aging mouse osteoblasts downregulate Spp1, contributing to the increased frequency of myeloid-biased HSCs, with compromised engraftment potential and loss of polarity.46 Therefore, we investigated the role of Spp1 in MDS progression by generating a chimeric model of an Spp1-KO mesenchymal environment, with hematopoietic cells of the MDS model, Vav-driven NHD13 fusion transgene. BM from NHD13 mice was competitively transplanted with WT BM. The NHD13 model recapitulates MDS through multilineage cytopenias and dysplasia, with evidence of a progressive and quantitative defect in HSPCs and high penetrance to transform to acute leukemias.26,47,48 Although the NHD13 genotype is uncommon in human MDS, the model was chosen because both the MDS and acute leukemia phases are well defined and stereotypic, providing effects of the ability to detect extrinsic cue on MDS progression.
Because of the age-dependent loss of the HSPC pool in NHD13 mice that is evidenced by a lower ability to engraft,47 we transplanted NHD13 (CD45-STEM) in competition with the WT CD45.2.STEM total BM at a ratio of 3:1 in lethally irradiated WT C57Bl6/J or Spp1-KO (CD45.2) mice and monitored the chimerism, MDS progression, and transformation through monthly peripheral blood analyses (Figure 4A). NHD13 donor cells demonstrated lower fitness than WT CD45.2.STEM cells in both WT and Spp1-KO recipients (Figure 4B-C). However, upon evaluating NHD13 donor cells in different recipient backgrounds, we found that chimerism was higher in Spp1-KO than in WT recipients (Figure 4D). This indicates an enhanced fitness of the NHD13 cells in the Spp1-KO microenvironment. The opposite was observed for WT hematopoietic cells. In that case, higher chimerism was noted when the donor cells were engrafted into WT recipients compared with Spp1-KO recipients (Figure 4E). The latter is in keeping with prior studies,46 in the effect of an Spp1-depleted microenvironment.
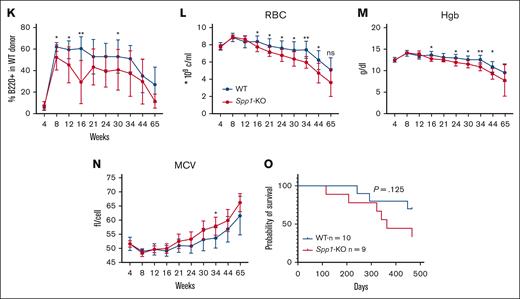
Accelerated progression of NHD13 MDS in Spp1-KO microenvironment. (A) A mix of NHD13STEM:WTCD45.2.STEM BM cells were transplanted in lethally irradiated CD45.2 WT (n = 10) or Spp1-KO (n = 9) mice at a ratio of 3:1. Mice were tracked via monthly peripheral blood analysis. (B-C) Chimerism analysis demonstrating a competitive advantage of WT over NHD13 donor cells in PB in both WT and Spp1-KO recipients. (D) Higher chimerism of NHD13 donor cells in Spp1-KO than in WT recipients. (E) Higher chimerism of WT donor cells in WT than in Spp1-KO recipients. (F) Higher contribution of NHD13 donor cells to peripheral blood myeloid compartment in Spp1-KO recipients. (G) Higher contribution of WT donor cells to peripheral blood myeloid compartment in WT recipients. (H) Very low contribution of NHD13 cells to donor CD3+ cells in WT and Spp1-KO recipients. (I) Competitor WT cells are a major source of donor derived CD3+ cells in WT and Spp1-KO recipients. (J) Very low contribution of NHD13 cells to donor B220+ cells in WT and Spp1-KO recipients. (K) Competitor WT cells are a major source of donor-derived B220+ cells in WT and Spp1-KO recipients. (L) Peripheral blood RBC count, (M) hemoglobin, and (N) mean corpuscular volume (MCV). (O) Survival analysis of WT and Spp1-KO animals that received NHD13 transplant. Data in panels B-N are presented as mean ± SD; ∗P < .05; ∗∗P < .01. Statistical significance was calculated using 2-way analysis of variance; multiple comparisons were corrected for using original FDR method of Benjamini and Hochberg.
Accelerated progression of NHD13 MDS in Spp1-KO microenvironment. (A) A mix of NHD13STEM:WTCD45.2.STEM BM cells were transplanted in lethally irradiated CD45.2 WT (n = 10) or Spp1-KO (n = 9) mice at a ratio of 3:1. Mice were tracked via monthly peripheral blood analysis. (B-C) Chimerism analysis demonstrating a competitive advantage of WT over NHD13 donor cells in PB in both WT and Spp1-KO recipients. (D) Higher chimerism of NHD13 donor cells in Spp1-KO than in WT recipients. (E) Higher chimerism of WT donor cells in WT than in Spp1-KO recipients. (F) Higher contribution of NHD13 donor cells to peripheral blood myeloid compartment in Spp1-KO recipients. (G) Higher contribution of WT donor cells to peripheral blood myeloid compartment in WT recipients. (H) Very low contribution of NHD13 cells to donor CD3+ cells in WT and Spp1-KO recipients. (I) Competitor WT cells are a major source of donor derived CD3+ cells in WT and Spp1-KO recipients. (J) Very low contribution of NHD13 cells to donor B220+ cells in WT and Spp1-KO recipients. (K) Competitor WT cells are a major source of donor-derived B220+ cells in WT and Spp1-KO recipients. (L) Peripheral blood RBC count, (M) hemoglobin, and (N) mean corpuscular volume (MCV). (O) Survival analysis of WT and Spp1-KO animals that received NHD13 transplant. Data in panels B-N are presented as mean ± SD; ∗P < .05; ∗∗P < .01. Statistical significance was calculated using 2-way analysis of variance; multiple comparisons were corrected for using original FDR method of Benjamini and Hochberg.
Donor-derived myeloid cells were mostly derived from NHD13 cells with higher contribution in Spp1-KO recipients (Figure 4F-G). In contrast, donor-derived lymphoid cells were mostly derived from WT donor cells (Figure 4H-K), indicating a myeloid skewing of the NHD13 cell that is more pronounced in the Spp1-KO recipients than in WT recipients. Furthermore, Spp1-KO recipient mice demonstrated evidence of anemia (Figure 4L-M) and a trend toward higher mean corpuscular volume (Figure 4N), which we interpret as incipient dysplasia. This was supported by a shorter latency of the disease in those animals, although only a modest trend was noted in survival (Figure 4O).
To rule out an immune-mediated effect for residual Spp1-KO hematopoietic cells in recipient mice, we transplanted NHD13 CD45-STEM cells in competition with WT or Spp1-KO CD45.2 total BM at a ratio of 3:1 in lethally irradiated WT heterozygous CD45.2.STEM mice (supplemental Figure 2A) and monitored the chimerism via monthly peripheral blood analysis. In this setting, the immune cells lacked the expression of Spp1 in 1 cohort, whereas the MSCs were WT in both. We hypothesized that if the MDS acceleration observed initially was due to the loss of expression of Spp1 in the nonhematopoietic stromal compartment only and not in the immune cells, then it should not be observed in this setting. Indeed, this was confirmed through the lack of significant difference in the contribution of NHD13 donor cells to the myeloid compartment, in which the competitor immune cells were devoid of Spp1 expression (supplemental Figure 2B-C), and no difference in the RBC parameters (supplemental Figure 2D-F).
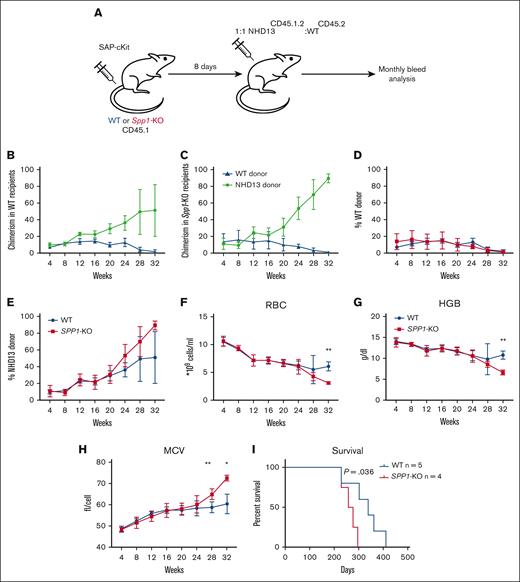
To further confirm that the enhanced fitness of NHD13 cells in the Spp1-KO microenvironment is not due to an irradiation-induced engraftment defect that favors NHD13 cells over WT, we repeated the experiment using the ADC as a conditioning strategy that spares the nonhematopoietic microenvironment in the BM from genotoxic injury.31 We chose to use the CD117-Saporin (ckit-SAP) conjugate that we generated using a biotinylated antibody and streptavidin-SAP conjugate as described previously.31,49 This approach has been shown to effectively deplete host HSCs with minimal toxicity.49 WT or Spp1-KO mice (CD45.1) were dosed with 3 mg/Kg ckit-SAP, and after 8 days, they were received transplants with 10 × 106 mix of NHD13(CD45.2.1):WT (CD45.2) total BM cells at a 1:1 ratio (Figure 5A). We deliberately chose to reduce the ratio of NHD13:WT cells in this experiment to rule out any advantage due to more NHD13 cells transplanted. Mice were monitored for engraftment and disease progression via monthly peripheral blood analysis up to 32 weeks. Unlike conditioning via irradiation, the NHD13 cells did not demonstrate lower fitness in the early weeks after transplantation (Figure 5B-C), with a trend toward higher chimerism than WT donor cells in Spp1-KO (Figure 5D-E). This confirms the enhanced fitness of NHD13 cells in the Spp1-KO microenvironment. By 32 weeks after transplant, the Spp1-KO recipients demonstrated evidence of anemia, reflected by reduced RBC counts and hemoglobin level and increased mean corpuscular volume (Figure 5F-H), with accelerated disease progression that resulted in a significant difference in survival (Figure 5I). Both Spp1-KO and WT recipients demonstrated enlarged spleens and elevated white blood cell counts at term (supplemental Figure 3A-D), indicating leukemic transformation.
Accelerated progression of NHD13 MDS in SPP1-KO microenvironment uninjured by irradiation. (A) A mix of NHD13CD45.2.1:WTCD45.2 BM cells were transplanted in lethally irradiated CD45.2 WT (n = 5) or Spp1-KO (n = 4) mice at a ratio of 1:1. Mice were tracked via monthly peripheral blood analysis. (B-C) Chimerism analysis of donor cells in WT and SPP1-KO recipients. (D-E) A trend of higher chimerism of NHD13 donor cells in Spp1-KO than in WT recipients. (F) Peripheral blood RBC count, (G) hemoglobin, and (H) MCV. (I) Survival analysis of WT and Spp1-KO animals that received NHD13 transplant. Data in panels F-H are presented as mean ± SD; ∗P < .05; ∗∗P < .01. Statistical analysis was calculated using multiple Student t test.
Accelerated progression of NHD13 MDS in SPP1-KO microenvironment uninjured by irradiation. (A) A mix of NHD13CD45.2.1:WTCD45.2 BM cells were transplanted in lethally irradiated CD45.2 WT (n = 5) or Spp1-KO (n = 4) mice at a ratio of 1:1. Mice were tracked via monthly peripheral blood analysis. (B-C) Chimerism analysis of donor cells in WT and SPP1-KO recipients. (D-E) A trend of higher chimerism of NHD13 donor cells in Spp1-KO than in WT recipients. (F) Peripheral blood RBC count, (G) hemoglobin, and (H) MCV. (I) Survival analysis of WT and Spp1-KO animals that received NHD13 transplant. Data in panels F-H are presented as mean ± SD; ∗P < .05; ∗∗P < .01. Statistical analysis was calculated using multiple Student t test.
Altogether, these findings confirm the enhanced fitness of NHD13 MDS in the context of a Spp1-devoid microenvironment, which is mediated by the nonhematopoietic stromal compartment.
Discussion
Transcriptomic and mutational analyses in combination with animal modeling have dramatically enhanced our understanding of MDS by identifying and functionally validating the role of point mutations in its pathophysiology.6,7,50,51 However, the therapeutics remain limited for MDS, in which the only curative therapy is HSC transplantation, a poor option for many of the older adult patients affected by the disease.52
The hypothesis that there may be marrow microenvironment abnormalities that contribute to MDS pathophysiology has gained experimental support, with stromal changes noted in the contexts of aging,46,53 inflammation,9 or genetic mutations in specific mesenchymal subsets in the BM.12-14,16
Here, we evaluated mesenchymal cell molecular features, seeking modifications that might affect MDS and offer therapeutic opportunities. We used comparative gene expression profiling to identify the human mesenchymal population most closely resembling the Osx-expressing cells defined to induce hematopoietic dysplasia in mice.13,14 Transcriptional differences of human mesenchymal cells expressing CD106 from other cell types were most similar to the differences between murine Osx-Cre– and Ocn-Cre–labeled cells. Gene expression profiling of 4 distinct mesenchymal subsets from patients with MDS and matched controls with benign disease demonstrated that, indeed, subsets labeled with CD106 in patients with MDS were the most distinct from the control subsets. It is worth noting that we could not detect significant differences in the frequency of these mesenchymal subsets.
GSEA demonstrated distinct signatures, with decreased osteogenic differentiation in the S subset and enrichment of inflammatory signatures in the DP and DN subsets, confirming prior findings,54,55 as well as an epithelial-to-mesenchymal transition signature. Within this signature, SPP1 was among the most highly upregulated genes and showed the highest number of interaction channels with MDS-propagating CD34+CD38− cells. Notably, mesenchyma-derived Spp1 was identified by us and others to be upregulated in the context of a murine model of AML,17,56 suggesting that this gene may be more broadly responsive to aberrant myelopoiesis. Spp1 is a negative regulator of the normal HSC number44,45 but protects their function,46 perhaps by preserving self-renewal. Through the competitive chimeric model of the NHD13 and WT BM, we demonstrated that an environment lacking Spp1 unexpectedly accelerates the progression of NHD13 dysplasia. This was evident in the increased chimerism of NHD13 donor cells, their increased myeloid contribution, and evidence of anemia in the Spp1-KO recipients. We confirmed that the residual recipient immune cells seen in the peripheral blood via flow cytometry did not contribute to the phenotype in the Spp1-KO mice.
Because increased SPP1 messenger RNA has been noted in cultured BM mesenchymal cells in patients with MDS and in animal models of AML,17,57 it may be that SPP1 reflects a generalized stromal response to aberrant myeloid stem or progenitors, perhaps in concert with the cytokine responses often observed. Here we show that it may be protective, reducing the fitness of NHD13-driven MDS. Rather than being an adverse signal to normal HSCs, it may serve to preserve WT HSC function while limiting the fitness of NHD13 mutant cells. The basis for this response is unclear, but Spp1 is known to be induced by inflammatory stimuli in some cell types.58
Altogether, our data identify molecular perturbations in human BM mesenchymal subsets that are specifically associated with MDS and that negatively affect pathogenesis in a mouse model of the disease. These results also suggest that niche changes are not restricted to those that lead to disease progression, as has been suggested by prior reports. Rather, we demonstrate that changes in SPP1 production may be protective and may serve to delay disease progression, ultimately protecting the host. MDS BM niche biology may, therefore, play a dual role, with certain modifications inducing or advancing disease and others providing protective adaptations.
Acknowledgments
The authors thank Maris Handley and the staff of the Harvard Stem Cell Institute-Center for Regenerative Medicine Flow Cytometry core for assistance with flow cytometry and cell sorting.
This study was supported by research funding from the Dubai Harvard foundation for medical research (Y.S.K.), Aplastic anemia and MDS International Foundation (Y.S.K.), STARR Cancer consortium (D.T.S. and O.A.-W.), Leukemia and Lymphoma Society (D.T.S.), and Gerald and Darlene Jordan Professorship (D.T.S.).
Authorship
Contribution: Y.S.K. designed the research, performed the experiments, analyzed data, and wrote the manuscript; F.J., and E.J. performed bioinformatic analysis; R.I.S. supervised bioinformatic analysis; G.S., A.P., F.M., M.M., and A.K. performed experiments; D.B.S., O.A.-W., M.R., and L.M.C. provided key reagents; and D.T.S. supervised the work, designed the research, and wrote the manuscript.
Conflict-of-interest disclosure: Y.S.K. is currently employed and holds equity at Moderna, Inc (unrelated to this manuscript). E.J. is an employee at Repare therapeutics (unrelated to this manuscript). G.S. is an employee at Tessera Therapeutics (unrelated to this manuscript). D.B.S. is a co-founder and holds equity in Clear Creek Bio (unrelated to this manuscript). O.A.-W. has served as a consultant for H3B Biomedicine, Foundation Medicine Inc, Merck, Prelude Therapeutics, and Janssen, and is on the scientific advisory board of Envisagenics Inc, AIChemy, Harmonic Discovery Inc, and Pfizer Boulder; and has received prior research funding from H3B Biomedicine, Loxo Oncology, and Nurix, unrelated to the current manuscript. D.T.S is founder and shareholder of Fate Therapeutics and Garuda Therapeutics, a founder, shareholder and director of Magenta Therapeutics, Lightning Biotherapeutics and Clear Creek Bio, a director and shareholder of Agios Pharmaceuticals, Editas Medicine and Sonata Therapeutics, a scientific advisory board member to Simcere Pharmaceuticals and VCanBio and previously received research support from Sumitomo Dianippon and Novartis, all unrelated to the current manuscript. The remaining authors declare no competing financial interests.
Correspondence: David T. Scadden, Center for Regenerative Medicine, Massachusetts General Hospital, 185 Cambridge St, Boston, MA 02114; e-mail: dscadden@mgh.harvard.edu; and Youmna S. Kfoury, Center for Regenerative Medicine, Massachusetts General Hospital, 185 Cambridge St, Boston, MA 02114; e-mail: youmna.kfoury@gmail.com.
References
Author notes
Sequencing data have been deposited in the Gene Expression Omnibus database (accession number GSE212639).
Data are available on request from the corresponding authors, David T. Scadden (dscadden@mgh.harvard.edu) and Youmna S. Kfoury (youmna.kfoury@gmail.com).
The full-text version of this article contains a data supplement.