Key Points
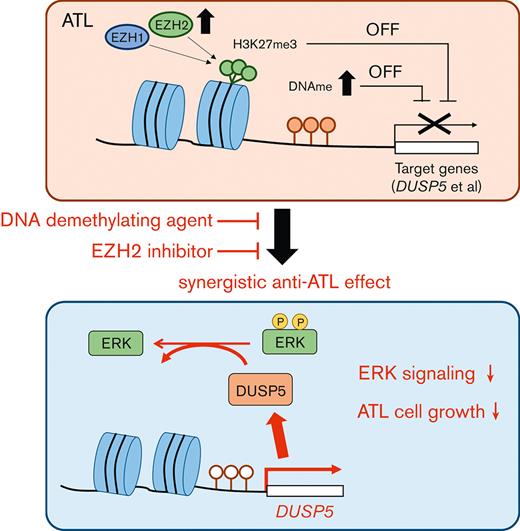
The combination of DNA demethylating agents and EZH inhibitors synergistically suppressed tumor cell growth in ATL.
DUSP5 is silenced in HTLV-1–infected cells via DNA and H3K27 methylation and the reactivation dephosphorylates ERK and inhibits cell growth.
Abstract
Adult T-cell leukemia/lymphoma (ATL) is a malignancy of mature CD4+ T cells caused by human T-cell lymphotropic virus type 1 (HTLV-1)–induced T-cell transformation. After infection with HTLV-1, it takes several decades for HTLV-1 carriers to develop ATL. The prognosis of ATL remains poor despite several new agents being approved in the last few years. Recently, it has been noted that epigenetic abnormalities, both DNA methylation and trimethylation at histone H3Lys27 (H3K27me3), contribute to ATL leukemogenesis. Here, we investigated the effect of combination treatment with DNA demethylating agents (azacitidine [AZA], decitabine (DAC), and OR-2100 (OR21), which is a silylated derivative of DAC) and inhibitors of enhancer of zeste homolog 2 (EZH2) (EPZ-6438 and DS-3201b), which catalyze trimethylation of H3K27, in ATL. The combination of DAC and OR21 but not AZA with EZH inhibitors exhibited synergistic anti-ATL effects in vitro and in vivo, concomitant with DNA demethylation and reduction of H3K27me3. The combination induced gene expression reprogramming. Dual-specificity phosphatase 5 (DUSP5), an extracellular signal-regulated kinase (ERK)–specific phosphatase, was identified as a key molecule that mediated the inhibitory effect of combination treatment by inactivating the ERK signaling pathway. DUSP5 was downregulated by DNA methylation and H3K27me3 accumulation in the promoter region in HTLV-1–infected cells from patients with ATL during ATL leukemogenesis. The present results demonstrate that dual targeting of aberrant DNA and histone methylation synergistically suppresses tumor cell growth by restoring DUSP5, and that dual targeting of aberrant DNA and histone methylation is a feasible therapeutic approach for ATL.
Introduction
Adult T-cell leukemia/lymphoma (ATL) is a highly malignant T-cell neoplasm caused by persistent infection with human T-lymphotropic virus 1 (HTLV-1).1 ATL is divided into 4 subtypes (acute, lymphoma, chronic, and smoldering), according to the Shimoyama classification.2 Indolent ATLs, such as smoldering and favorable chronic types, are treated by watchful waiting3 in Japan, whereas various intensive chemotherapies followed by allogeneic hematopoietic stem cell transplantation are used for the management of aggressive ATLs, such as acute, lymphoma, and unfavorable chronic types.4,5 Despite the available treatments, the prognosis of aggressive ATL remains poor, with a median survival time of ∼1 year, and that of indolent ATL has not improved in 30 years.2,6 Recently, an anti-CCR4 antibody (mogamulizumab),7 an immunomodulatory agent (lenalidomide),8 and an inhibitor of histone deacetylase (tucidinostat)9 were approved for the treatment of aggressive ATL. However, most patients with ATL treated with mogamulizumab relapsed or became refractory after a short period,10 underscoring the need to identify effective therapeutic strategies for ATL.
HTLV-1–infected T cells are transformed into aggressive ATL cells through a multistep leukemogenesis process characterized by the accumulation of genetic11,12 and epigenetic abnormalities.11,13-15 Tax, an HTLV-1 oncoprotein, activates the promoter of enhancer of zeste homolog 2 (EZH2), which catalyzes the trimethylation of histone H3 at Lys 27 (H3K27me3),16 and abnormal H3K27me3 accumulation occurs during the progression of ATL.17-20 EZH1, a homolog that catalyzes the formation of H3K27me3,21 is expressed at high levels in normal CD4+ T cells and ATL cells.20 EZH2 inhibitors and an EZH1/EZH2 dual inhibitor show anti-ATL effects in preclinical studies,19,20 and their effects in non-Hodgkin lymphomas, including ATL, are being tested in clinical trials.22 ATL-specific DNA hypermethylation has also been reported,11 and the expression of ATL-related genes is decreased by promoter region DNA hypermethylation.13,23 We previously reported that regional DNA hypermethylation reflects the ATL disease status and that DNA demethylating agents, such as azacitidine (AZA), decitabine (DAC), and OR-2100 (OR21), exert anti-ATL effects by restoring gene expression.24 OR21 is a silylated derivative of DAC that is resistant to cytidine deaminase and can be administered orally.24-26 These previous results led us to investigate the effect of combination treatment with DNA demethylating agents and EZH2 inhibitors in patients with ATL.
Here, we found that DNA demethylating agents and EZH2 inhibitors had synergistic activity against ATL and restored dual-specificity phosphatase 5 (DUSP5) expression. DUSP5 is a member of the DUSP family27 that specifically dephosphorylates extracellular signal-regulated kinase (ERK) and anchors inactive ERK in the nucleus,28,29 in addition to its function as a tumor suppressor in skin and colorectal cancer.28,30 Therefore, we also investigated the expression level and the functional role of DUSP5 in ATL leukemogenesis.
Materials and methods
Human samples
All studies using human samples were performed in accordance with the guidelines of the Declaration of Helsinki and approved by the institutional review board of Saga University (2018-03-02). Primary peripheral blood mononuclear cells (PBMCs) from patients with ATL, asymptomatic HTLV-1 carriers, and healthy volunteers (supplemental Table 1) were isolated by Ficoll separation as described previously.24 All HTLV-1 carriers, patients with ATL, and healthy volunteers provided written informed consent.
HTLV-1–infected human T-cell lines
Seven different HTLV-1–infected human T-cell lines were used. MT-1, MT-2, and MT-4 were from the JCRB Cell Bank; ATN-1 and TL-Mor were from the RIKEN Bio Resource Center; MJ was from the American Type Culture Collection; and TL-Om1 was provided by Masao Matsuoka (Kumamoto University). Cell lines were cultured as we previously reported.24
Reagents
AZA and DAC were purchased from Sigma-Aldrich. OR21 was from OHARA Pharmaceutical Co (Shiga, Japan). EPZ-6438 (EPZ) and DS-3201b (DS) were purchased from Active Biochem (Kowloon, Hong Kong) and Chemietek (Indianapolis, IN), respectively. All reagents were dissolved in dimethyl sulfoxide and stored at −20°C.
Cell growth assay
HTLV-1–infected human T-cell lines were cultured in the presence of the indicated compounds, which were added at 0, 48, and 96 hours after seeding. Cell growth was measured using a Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technology). The 50% inhibitory concentration values were calculated for each compound as described previously.31
Synergy determination with the SynergyFinder
MT-1 and MT-2 cells were treated with 5 concentrations of DNA demethylating agents (AZA, DAC, or OR21) and 5 concentrations of EPZ-6438 for 4 days (5 × 5, total 25 conditions). Each compound was added every 48 hours. The synergy score was determined using the “inhibition readout” (calculated relative to the control as 100%) on the online SynergyFinder software32 and implementing the zero interaction potency (ZIP) calculation method.33
Bisulfite pyrosequencing
The methylation status of CpG sites of LINE-1 and DUSP5 was determined using pyrosequencing-based analysis as described in detail in supplemental Methods.
Quantitative real-time polymerase chain reaction (PCR)
Cells were lysed in the TRIzol reagent (Invitrogen), and total RNA was purified using Direct-zol RNA MiniPrep Kits (Zymo Research). Complementary DNA (cDNA) was produced using ReverTra Ace (TOYOBO), and quantitative real-time PCR was performed using THUNDERBIRD SYBR quantitative PCR Mix (TOYOBO). The gene expression level of ACTB was used as an internal control. The oligonucleotides are listed in supplemental Methods.
Gene expression microarray analysis
DNaseI-treated total RNAs were prepared using the Direct-zol RNA MiniPrep Kit (Zymo Research). cDNA was labeled using the GeneChip WT PLUS Reagent Kit (Thermo Fisher Scientific). Array scanning was performed using the GeneChip Scanner 3000 7G (Thermo Fisher Scientific), and whole gene expression data were obtained (Filgen). Data analysis was performed using Microarray Data Analysis Tool Ver3.2 (Filgen).
Genome-wide DNA methylation analysis
Genomic DNA was extracted from MT-1 cells using a QIAamp DNA Mini Kit (Qiagen). Genome-wide DNA methylation analysis was performed using Infinium Human Methylation EPIC Bead Chips (Illumina) as described previously.26 β-values ranged from 0 (unmethylated) to 1 (fully methylated). To reduce the data size and obtain data that were easy to handle, the CpG probes were grouped into 551 478 genomic blocks (GBs), which consisted of probes within 500 bp.34
Histone extraction
Histone extraction was performed as described previously.35 Histone proteins were extracted in water, and the total histone protein content was quantified using a protein assay (Bio-Rad). Equal amounts of each histone protein were used for western blot analysis.
Flow cytometry and sorting
Primary PBMCs were stained with anti-CD3, anti-CD4, anti-CD7, and anti-CADM1 antibodies at room temperature in phosphate-buffered saline (−) (Wako) containing 2% fetal bovine serum and sorted into CD7+CADM1− cells and CADM1+ cells on a FACSAriaII (BD Biosciences). Flow cytometry data were analyzed using FlowJo software (Becton Dickinson & Company). The antibodies are listed in supplemental Methods.
Western blot analysis
Whole cell lysates were extracted using RIPA buffer (Santa Cruz Biotechnology). The total protein content was quantified using a protein assay (Bio-Rad). Each lysate was resolved in n-polyacrylamide gels (Invitrogen), and the proteins were transferred onto nitrocellulose membranes. Each protein was detected using an ECL detection reagent (GE Healthcare). The antibodies are listed in supplemental Methods.
Lentivirus preparation and infection
Lentiviral particles for transduction of DUSP5 cDNA (VB200115-1034cym, pLV[Exp]-Neo-EF1A.hDUSP5[NM_004419]:IRES:EGFP) and control GFP/mCherry lentiviral particles (VB160109-10005, pLV[Exp]-EGFP:T2A:Puro-EF1A.mCherry) were prepared by VectorBuilder. HTLV-1–infected cell lines were infected by lentiviral particles using RetroNectin (Takara).
ATL cell xenograft mouse model
Animal studies were conducted in accordance with Saga University–approved animal protocols at Saga University (G2020-09-01) and the German Animal Welfare Act. NOD/Shi-scid, IL-2Rγ KO Jic (NOG) female mice, 6 weeks of age, were obtained from In-Vivo Science Inc (Kawasaki, Japan). NOG mice subcutaneously inoculated with TL-Om1 cells were treated with vehicle, OR21, EPZ, and a combination of OR21 and EPZ. Details are described in supplemental Methods.
Statistics
Data are expressed as the mean ± standard deviation. Differences between groups were tested using the Student t test, Mann-Whitney U test, Kruskal-Wallis test, or 1-way analysis of variance. Calculations were performed using R v.3.4.4 (The R Foundation for Statistical Computing, Vienna, Austria) or EZR36 (a graphical user interface for R; Saitama Medical Center, Jichi Medical University, Saitama, Japan). Differences between samples from the same volunteer were tested using the paired t test. This calculation was performed using Microsoft Excel software (Microsoft Corp, Seattle, WA). The results were considered significant when a P value <.05 was obtained by 2-sided tests.
Public database analyses
Microarray data from the Gene Expression Omnibus (GEO) under accession number GSE33615 were analyzed to measure gene expression in healthy volunteers and patients with ATL.17 DNA methylation data from GEO under accession number GSE136189, which we previously reported, were analyzed to measure the β-value of patients with ATL and ATL cell lines.24 Chromatin immunoprecipitation on-chip data from the GEO database under accession number GSE71450 were analyzed to measure the levels of H3K27me3 in patients with ATL and ATL cell lines.19
Results
Anti-ATL effects of combination treatment with DNA demethylating agents and EZH2 inhibitors
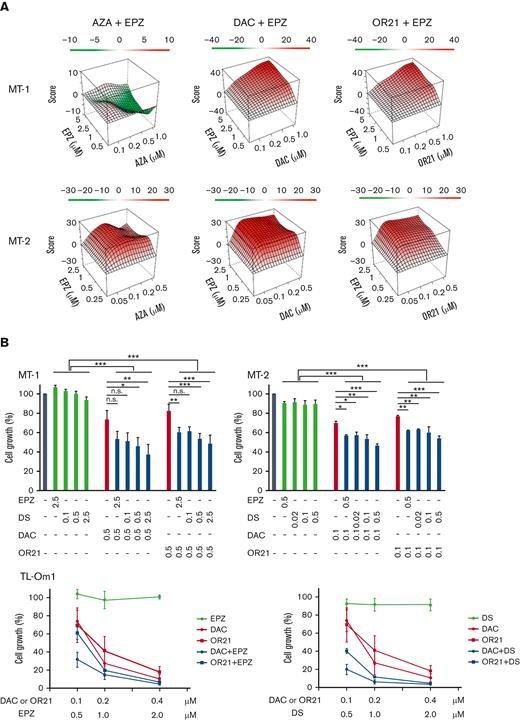
The growth inhibitory effects of EPZ-6438 (EPZ, EZH2 inhibitor) and DS-3201b (DS, EZH1/2 dual inhibitor) in HTLV-1–infected cell lines or ATL cell lines (MT-1, MT-2, TL-Om1, and TL-Mor) were measured using the CCK-8 assay. MT-1 and MT-2 cells were less sensitive to EPZ and DS monotherapies than TL-Om1 and TL-Mor cells (supplemental Figure 1). We previously reported that MT-1 cells are less sensitive to DAC and OR21 than other ATL cells.24 Therefore, we assessed the effects of combination treatment with DNA demethylating agents (AZA, DAC, or OR21) and an EZH2 inhibitor (EPZ) in MT-1 and MT-2 cells. DAC or OR21 showed synergistic effects with EPZ in both MT-1 and MT-2 cells (Figure 1A; supplemental Table 2). The combination of AZA and EPZ did not show a synergistic effect in MT-1 cells (Figure 1A; supplemental Table 2). The combination of DAC or OR21 with DS at lower concentrations showed a growth inhibitory effect comparable to the combination of DAC or OR21 with EPZ (Figure 1B, upper panels). Although TL-Om1 cells are sensitive to EZH2 inhibitors, a long exposure of 1 to 2 weeks is necessary for efficacy.19,20 Consistently, we found that EPZ inhibitors alone had no growth inhibitory effects at 4 days, even at high concentrations (Figure 1B, lower panels). However, combination treatment for 4 days inhibited cell growth even at lower concentrations (Figure 1B, lower panels).
Inhibition of ATL cell growth by DNA demethylating agents and EZH2 inhibitors in vitro. (A) MT-1 and MT-2 cells were treated with each compound alone or in combination for 4 days, and cell growth was assessed using the CCK-8 reagent. Synergy scores were calculated using the SynergyFinder software.32 ZIP Synergy scores −10 to 10 indicate additive, >10 indicate synergistic, and less than −10 indicate antagonistic effects.33 (B) MT-1, MT-2, and TL-Om1 cells were cultured for 4 days with DNA demethylating agents (DAC or OR21) in the presence or absence of EZH2 inhibitors (EPZ or DS). Cell growth was assessed using the CCK-8 reagent. The absorbance of untreated cells was defined as 100%. The results are expressed as the mean of 3 independent experiments ± SD. Differences between groups were tested using the Tukey test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. n.s., not significant; SD, standard deviation.
Inhibition of ATL cell growth by DNA demethylating agents and EZH2 inhibitors in vitro. (A) MT-1 and MT-2 cells were treated with each compound alone or in combination for 4 days, and cell growth was assessed using the CCK-8 reagent. Synergy scores were calculated using the SynergyFinder software.32 ZIP Synergy scores −10 to 10 indicate additive, >10 indicate synergistic, and less than −10 indicate antagonistic effects.33 (B) MT-1, MT-2, and TL-Om1 cells were cultured for 4 days with DNA demethylating agents (DAC or OR21) in the presence or absence of EZH2 inhibitors (EPZ or DS). Cell growth was assessed using the CCK-8 reagent. The absorbance of untreated cells was defined as 100%. The results are expressed as the mean of 3 independent experiments ± SD. Differences between groups were tested using the Tukey test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. n.s., not significant; SD, standard deviation.
Combination treatment with a DNA demethylating agent and an EZH2 inhibitor alters the gene expression profile
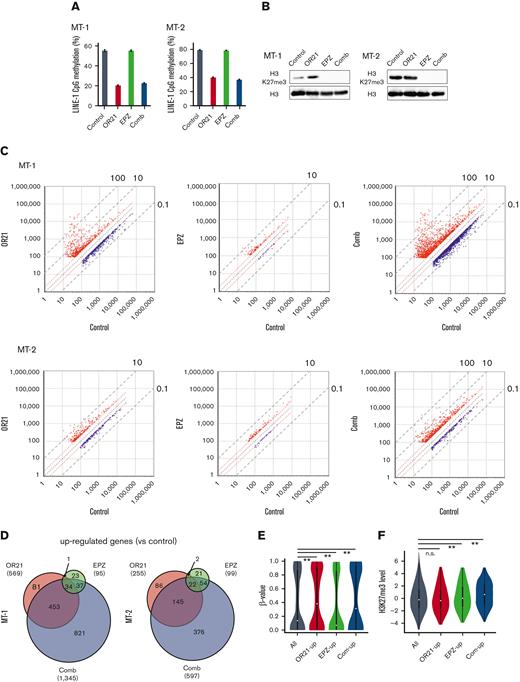
Abnormal DNA hypermethylation and the accumulation of H3K27me3 contribute to the downregulation of gene expression.19,20,24 We compared gene expression, DNA methylation, and H3K27me3 in normal CD4+ T cells and ATL cells using 3 data sets (GSE33615, GSE136189, and GSE71450) to count the number of genes that were downregulated concomitantly with DNA methylation and/or H3K27me3 in ATL. The expression of 12 452 genes were downregulated; DNA hypermethylation and H3K27 hypermethylation were found in 2474 genes and 6584 genes, respectively (supplemental Figure 2A). Among them, 1086 genes showed both abnormalities and their expressions were downregulated compared with genes that exhibited only DNA or H3K27 hypermethylation (supplemental Figure 2B). We next investigated the effects of combination on DNA methylation, H3K27me3, and gene expression. Long interspersed nuclear element-1 (LINE-1) constitutes ∼17% of human genomic DNA37; therefore, the LINE-1 methylation status is an indicator of the global DNA methylation status.38 As expected, OR21 monotherapy and EPZ monotherapy reduced LINE-1 methylation and H3K27me3, respectively, and the combination reduced both methylation (Figure 2A-B). Microarray analysis of gene expression profiles showed that, in both MT-1 and MT-2 cells, combination treatment had a greater effect on gene expression than each single treatment (Figure 2C-D; supplemental Figure 3). The combination largely upregulated the expression of genes that were downregulated by both DNA methylation and H3K27me3 compared with those of genes that are downregulated only by DNA methylation or H3K27me3 (supplemental Figure 2C). We measured DNA methylation and H3K27me3 levels in the group of genes upregulated by OR21 (OR21-up, 231 genes in Figure 2D right panel) or EPZ (EPZ-up, 75 genes in Figure 2D right panel) treatment and in those commonly upregulated by both OR21 and EPZ (com-up, 24 genes in Figure 2D right panel) in MT-2 cells. The DNA methylation level in TSS200 or TSS1500 (within 200 or 200-1500 bp upstream of the transcription start site) was higher in the OR21-up and com-up gene groups than in the whole gene group, whereas that of the EPZ-up gene group was not (Figure 2E). The H3K27me3 levels were higher in the promoter regions of the EPZ-up and com-up gene groups than in those of the whole gene group, whereas those of the OR21-up group were not (Figure 2F). These results indicate that genes upregulated by both OR21 and EPZ showed DNA and H3K27 hypermethylation in their promoter regions, suggesting that both DNA demethylation and reduction of H3K27me3 contributed to the modulation of gene expression.
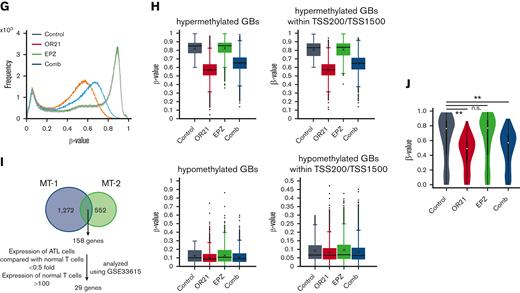
Global epigenetic changes associated with gene expression reprogramming induced by the combination of OR21 and EPZ-6438. MT-1 and MT-2 cells were treated with OR21, EPZ, or OR21 + EPZ (Comb) for 4 days (MT-1: 0.5 μM OR21 and/or 2.5 μM EPZ; MT-2: 0.1 μM OR21 and/or 0.5 μM EPZ). (A) The LINE-1 promoter methylation level was determined using bisulfite pyrosequencing. Data are expressed as the means of 3 independent experiments ± SD. (B) Amounts of H3K27me3 were determined by immunoblotting. (C) Scatter plot showing upregulated (≥twofold) and downregulated genes (≤0.5-fold) (red and blue dots, respectively) compared with the control in global gene expression analysis using microarrays. (D) Venn diagrams showing the number of upregulated genes. (E) β-values in the TSS200 or TSS1500 region in MT-2 cells determined by global methylation analysis using GSE136189. The gray plots show β-values of all genes (n = 125 523); the red plots show β-values of OR21-up genes (n = 2536); the green plots show β-values of EPZ-up genes (n = 786); the blue plots show β-values of com-up genes (n = 180). OR21-up genes are genes upregulated by OR21 monotherapy in MT-2 cells in panel D. EPZ-up genes are genes upregulated by EPZ monotherapy in MT-2 cells in panel D. Com-up genes are genes commonly upregulated by both OR21 and EPZ in MT-2 cells in panel D. Differences between all and each group were tested using Steel test. ∗∗P < .01. (F) H3K27me3 levels (log2) around the promoter region in MT-2 cells obtained from public data (GSE71450). The gray (n = 202 178), red (n = 1759), green (n = 711), and blue (n = 186) plots indicate the same gene groups shown in panel E. Differences between all and each group were tested using Steel test. ∗∗P < .01. (G) Histogram showing the frequency of β-values under each condition (control, OR21, EPZ, Comb) in all GBs in MT-1 cells. (H) Box plots showing the β-values under each condition (control, OR21, EPZ, Comb). Left panels: β-values of hypermethylated GBs (β-value in the control condition >0.6, upper panel) and hypomethylated GBs (β-value in the control condition <0.3, lower panel). Right panels: β-values of hypermethylated (upper panel) or hypomethylated (lower panel) GBs within TSS200 and TSS1500 region in MT-1 cells. (I) Venn diagrams showing the number of upregulated genes increased by combination treatment compared with single treatment; 158 genes were common to MT-1 and MT-2 cells. Twenty-nine genes were expressed in normal T cells and downregulated in cells from patients with ATL compared with normal T cells (analyzed using GSE33615) were selected as candidate genes involved in the synergistic anti-ATL effects. (J) β-values in the TSS200 or TSS1500 region of 29 genes after OR21 and/or EPZ treatment in MT-1 cells. Differences between control and each condition were tested using Steel test. ∗∗P < .01. n.s., not significant; SD, standard deviation.
Global epigenetic changes associated with gene expression reprogramming induced by the combination of OR21 and EPZ-6438. MT-1 and MT-2 cells were treated with OR21, EPZ, or OR21 + EPZ (Comb) for 4 days (MT-1: 0.5 μM OR21 and/or 2.5 μM EPZ; MT-2: 0.1 μM OR21 and/or 0.5 μM EPZ). (A) The LINE-1 promoter methylation level was determined using bisulfite pyrosequencing. Data are expressed as the means of 3 independent experiments ± SD. (B) Amounts of H3K27me3 were determined by immunoblotting. (C) Scatter plot showing upregulated (≥twofold) and downregulated genes (≤0.5-fold) (red and blue dots, respectively) compared with the control in global gene expression analysis using microarrays. (D) Venn diagrams showing the number of upregulated genes. (E) β-values in the TSS200 or TSS1500 region in MT-2 cells determined by global methylation analysis using GSE136189. The gray plots show β-values of all genes (n = 125 523); the red plots show β-values of OR21-up genes (n = 2536); the green plots show β-values of EPZ-up genes (n = 786); the blue plots show β-values of com-up genes (n = 180). OR21-up genes are genes upregulated by OR21 monotherapy in MT-2 cells in panel D. EPZ-up genes are genes upregulated by EPZ monotherapy in MT-2 cells in panel D. Com-up genes are genes commonly upregulated by both OR21 and EPZ in MT-2 cells in panel D. Differences between all and each group were tested using Steel test. ∗∗P < .01. (F) H3K27me3 levels (log2) around the promoter region in MT-2 cells obtained from public data (GSE71450). The gray (n = 202 178), red (n = 1759), green (n = 711), and blue (n = 186) plots indicate the same gene groups shown in panel E. Differences between all and each group were tested using Steel test. ∗∗P < .01. (G) Histogram showing the frequency of β-values under each condition (control, OR21, EPZ, Comb) in all GBs in MT-1 cells. (H) Box plots showing the β-values under each condition (control, OR21, EPZ, Comb). Left panels: β-values of hypermethylated GBs (β-value in the control condition >0.6, upper panel) and hypomethylated GBs (β-value in the control condition <0.3, lower panel). Right panels: β-values of hypermethylated (upper panel) or hypomethylated (lower panel) GBs within TSS200 and TSS1500 region in MT-1 cells. (I) Venn diagrams showing the number of upregulated genes increased by combination treatment compared with single treatment; 158 genes were common to MT-1 and MT-2 cells. Twenty-nine genes were expressed in normal T cells and downregulated in cells from patients with ATL compared with normal T cells (analyzed using GSE33615) were selected as candidate genes involved in the synergistic anti-ATL effects. (J) β-values in the TSS200 or TSS1500 region of 29 genes after OR21 and/or EPZ treatment in MT-1 cells. Differences between control and each condition were tested using Steel test. ∗∗P < .01. n.s., not significant; SD, standard deviation.
OR21 treatment alters the DNA methylation profile
As shown in Figure 2A-B, although the H3K27me3 signal was completely abolished in EPZ-treated groups, LINE1 methylation was partially reduced in OR21-treated groups. Therefore, we analyzed the demethylated GBs induced by treatment with OR21. Exposure to OR21 and combination treatment but not to EPZ alone caused global DNA hypomethylation (Figure 2G). The β-value of the combination was slightly higher than that of OR21 alone (Figure 2G). Analysis of hypermethylated GBs (β-value >0.6 in the control condition) and hypomethylated GBs (β-value <0.3 in the control condition) showed that the demethylation effects of OR21 and combination treatment were higher in hypermethylated GBs than in hypomethylated GBs (Figure 2H). To identify the genomic regions contributing to the increase in gene expression caused by combination treatment, we focused on genes upregulated by the combination and downregulated in cells from patients with ATL compared with normal T cells (29 genes in Figure 2I; supplemental Table 3). Of the 158 genes upregulated by combination treatment compared with single treatment in both MT-1 and MT-2 cells, 29 genes were downregulated in cells from patients with ATL compared with normal T cells (Figure 2I; supplemental Table 3). OR21 (with or without EPZ) decreased the promoter methylation of 29 genes (Figure 2J). These results indicate that OR21 demethylated the promoter regions of genes upregulated by combination treatment regardless of the presence of EPZ.
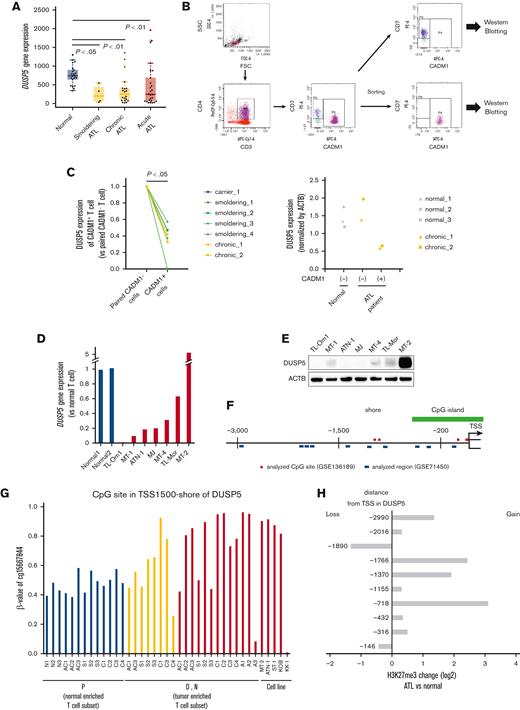
DUSP5 is downregulated in HTLV-1–infected cells during ATL leukemogenesis
The 29 genes identified in Figure 2I or supplemental Table 3 were further analyzed to identify the genes related to the synergistic anti-ATL effect of OR21 and EPZ. The group of 29 genes included 5 tumor suppressor genes (DUSP5, RASSF2, SRC, HPGD, and ERRFI1) registered in the tumor suppressor gene database. The 5 tumor suppressor genes were downregulated during ATL leukemogenesis (Figure 3A; supplemental Figure 4).
DUSP5 expression is silenced in HTLV-1–infected T cells during ATL leukemogenesis. (A) DUSP5 expression in healthy volunteers and patients with ATL was analyzed using GSE33615. Normal CD4+ T cells were isolated from healthy volunteers (n = 21). PBMCs were isolated from patients with smoldering (n = 4), chronic (n = 20), and acute (n = 26) ATL. Differences between normal and each ATL subtype were tested using Steel test. (B-C) HTLV-1–infected T cells (CADM1+ T cells) and normal counterparts (CD7+CADM1− T cells) were isolated from healthy volunteers (n = 3), HTLV-1 carriers (n = 1), and patients with smoldering ATL (n = 4) and chronic ATL (n = 2). DUSP5 expression was measured by western blotting and normalized to β-actin. Left panel: the ratio of DUSP5 expression in CADM1+ cells compared with paired CADM1− cells. Differences between each cell were tested using the paired t test. (D) DUSP5 expression was compared between HTLV-1–infected cell lines and CD3+CD4+CD7+CADM1− T cells (normal T cells) from 2 healthy volunteers. (E) DUSP5 protein expression in HTLV-1–infected cell lines was determined by western blotting. (F) Location of CpG sites analyzed using GSE136189 (red line) and H3K27me3 peaks analyzed using GSE71450 (blue line) around the DUSP5 promoter region. The green line indicates CpG islands. (G) β-values of TSS1500-shore CpG sites of DUSP5 in CD7+CADM1− T cells (subpopulation P), CD7+CADM1+ T cells (subpopulation D), and CD7−CADM1+ T cells (subpopulation N) of healthy volunteers, HTLV-1 carriers, patients with ATL, and ATL cell lines. The site was located in TSS1500-shore, as shown in supplemental Table 4. (H) H3K27me3 alternations of the 10 promoter regions in DUSP5 in patients with ATL (n = 3) compared with healthy volunteers (n = 2). The numbers of y-axis indicate distances from TSS, as shown in supplemental Table 5.
DUSP5 expression is silenced in HTLV-1–infected T cells during ATL leukemogenesis. (A) DUSP5 expression in healthy volunteers and patients with ATL was analyzed using GSE33615. Normal CD4+ T cells were isolated from healthy volunteers (n = 21). PBMCs were isolated from patients with smoldering (n = 4), chronic (n = 20), and acute (n = 26) ATL. Differences between normal and each ATL subtype were tested using Steel test. (B-C) HTLV-1–infected T cells (CADM1+ T cells) and normal counterparts (CD7+CADM1− T cells) were isolated from healthy volunteers (n = 3), HTLV-1 carriers (n = 1), and patients with smoldering ATL (n = 4) and chronic ATL (n = 2). DUSP5 expression was measured by western blotting and normalized to β-actin. Left panel: the ratio of DUSP5 expression in CADM1+ cells compared with paired CADM1− cells. Differences between each cell were tested using the paired t test. (D) DUSP5 expression was compared between HTLV-1–infected cell lines and CD3+CD4+CD7+CADM1− T cells (normal T cells) from 2 healthy volunteers. (E) DUSP5 protein expression in HTLV-1–infected cell lines was determined by western blotting. (F) Location of CpG sites analyzed using GSE136189 (red line) and H3K27me3 peaks analyzed using GSE71450 (blue line) around the DUSP5 promoter region. The green line indicates CpG islands. (G) β-values of TSS1500-shore CpG sites of DUSP5 in CD7+CADM1− T cells (subpopulation P), CD7+CADM1+ T cells (subpopulation D), and CD7−CADM1+ T cells (subpopulation N) of healthy volunteers, HTLV-1 carriers, patients with ATL, and ATL cell lines. The site was located in TSS1500-shore, as shown in supplemental Table 4. (H) H3K27me3 alternations of the 10 promoter regions in DUSP5 in patients with ATL (n = 3) compared with healthy volunteers (n = 2). The numbers of y-axis indicate distances from TSS, as shown in supplemental Table 5.
DUSP5 is expressed in CD4+ T cells,39 and DUSP5-expressing mature T cells exhibit decreased IL-2–dependent proliferation and IL-2–induced ERK1/2 phosphorylation in mice.40 We thus focused on DUSP5 as a potential mediator of the synergistic anti-ATL effects of combination treatment. The CADM1 vs CD7 plot accurately reflects ATL progression,24,41 and CADM1+ cells, which represent the HTLV-1–infected cell population, exhibit abnormal DNA hypermethylation and accumulation of H3K27me3.19,20,24 Therefore, we compared DUSP5 expression between CADM1− and CADM1+ T cells from the same patient. PBMCs from healthy volunteers and patients with ATL were fractionated into CD7+CADM1− and CADM1+ cells (Figure 3B). DUSP5 expression was significantly lower in CADM1+ T cells from a HTLV-1 carrier and patients with ATL than in CADM1− T cells (Figure 3C left panel; supplemental Figure 5). DUSP5 expression was comparable between CADM1− cells from patients with ATL and CADM1− cells from healthy volunteers (Figure 3C right panel; supplemental Figure 5). DUSP5 gene expression was lower in most ATL cell lines than in CADM1− T cells from healthy volunteers (Figure 3D). The DUSP5 protein was not detected or only slightly expressed in most ATL cell lines except in MT-2 cells (Figure 3E). To investigate CpG methylation and H3K27me3 of the DUSP5 promoter region in patients with ATL, HumanMethylation BeadChip data (GSE136189) and chip-on-chip data (GSE71450) were analyzed. The CpG sites of the TSS200-island and TSS1500-shore, and H3K27me3 of 10 regions around the promoter were investigated (Figure 3F). In the TSS1500-shore, the CpG sites of CD7+CADM1+ T cells (D) and CD7-CADM1+ T cells (N) tended to be hypermethylated compared with those of CD7+CADM1− T cells (P) (Figure 3G). In contrast, CpG sites around the TSS200-island were unmethylated in all specimens (supplemental Figure 6A). Most of the H3K27me3 levels at the DUSP5 promoter region were higher in patients with acute ATL than in healthy volunteers (Figure 3H). These results indicate that DUSP5 expression may be silenced by DNA hypermethylation and H3K27me3 accumulation within the DUSP5 promoter region in HTLV-1–infected T cells of patients with ATL.
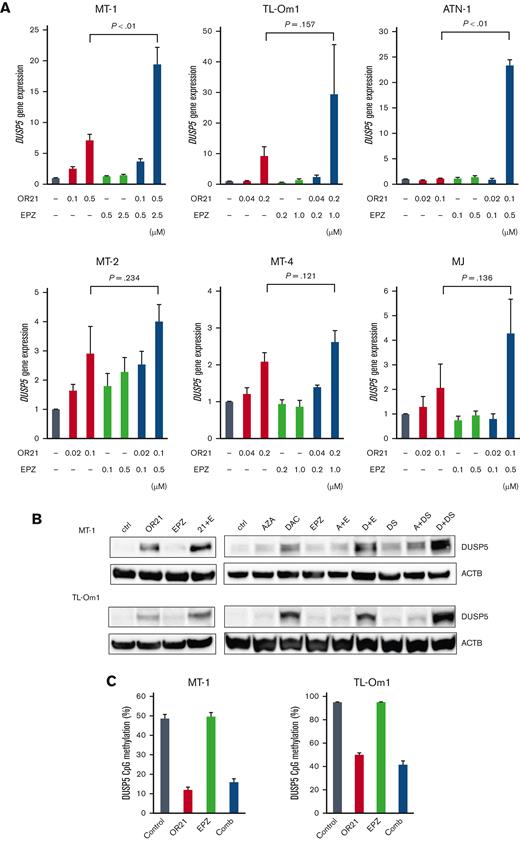
DUSP5 is upregulated by combination treatment with DNA demethylating agents and EZH2 inhibitors
Combination treatment upregulated DUSP5 expression in all cell lines (Figure 4A) and markedly increased DUSP5 protein expression (Figure 4B), as expected. The combination of DAC and DS had the greatest effect on inducing DUSP5 expression (Figure 4B), whereas the combination of AZA and EPZ did not upregulate DUSP5 expression compared with single treatment in MT-1 cells (Figure 4B). The effect of OR21 on the promoter DNA methylation of DUSP5 was investigated using pyrosequencing assays. OR21 (with or without EPZ) decreased the CpG methylation of DUSP5 in the TSS1500-shore (Figure 4C), but not in the TSS200-island (supplemental Figure 6B).
Induction of DUSP5 by the combination of DNA demethylating agents and EZH2 inhibitors. (A) DUSP5 gene expression in HTLV-1–infected cell lines and ATL cell lines treated with the indicated compounds. Differences were tested using the Student t test. (B) DUSP5 protein amounts were determined by western blotting after treatment with the indicated compounds: 0.5 μM OR21, 2.5 μM EPZ, 0.5 μM AZA, 0.5 μM DAC, and 0.25 μM DS for MT-1; 0.1 μM OR21, 0.5 μM EPZ, 0.1 μM AZA, 0.1 μM DAC, and 0.05 μM DS for TL-Om1 cells. (C) DNA methylation levels around the DUSP5 promoter region (at TSS1500-shore as indicated in Figure 3G) of cells treated with OR21 and/or EPZ.
Induction of DUSP5 by the combination of DNA demethylating agents and EZH2 inhibitors. (A) DUSP5 gene expression in HTLV-1–infected cell lines and ATL cell lines treated with the indicated compounds. Differences were tested using the Student t test. (B) DUSP5 protein amounts were determined by western blotting after treatment with the indicated compounds: 0.5 μM OR21, 2.5 μM EPZ, 0.5 μM AZA, 0.5 μM DAC, and 0.25 μM DS for MT-1; 0.1 μM OR21, 0.5 μM EPZ, 0.1 μM AZA, 0.1 μM DAC, and 0.05 μM DS for TL-Om1 cells. (C) DNA methylation levels around the DUSP5 promoter region (at TSS1500-shore as indicated in Figure 3G) of cells treated with OR21 and/or EPZ.
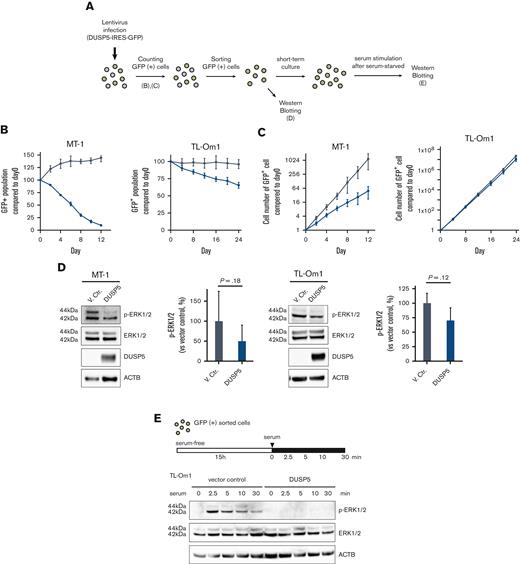
DUSP5 suppresses cell growth concomitant with dephosphorylation of ERK in ATL cells
We next examined whether induction of DUSP5 contributes to the anti-ATL effects of combination treatment. The MT-1 and TL-Om1 cell lines express small amounts of DUSP5 (Figure 3D-E), and thus were considered the most suitable lines for the rescue experiments. The 2 cell lines were transduced with lentiviral vectors expressing the DUSP5 and GFP genes (Figure 5A). The GFP+ population, which corresponded to DUSP5-expressing cells, decreased in a time-dependent manner, whereas cells transduced with the vector control did not (Figure 5B; supplemental Figure 7). Cell proliferation was also suppressed in DUSP5-expressing cells (Figure 5C). To confirm the function of DUSP5 as a specific phosphatase for ERK1/2, GFP+ cells were subjected to western blot analysis, which showed that exogenous DUSP5 expression tended to cause p-ERK1/2 dephosphorylation (Figure 5D). ERK1/2 phosphorylation was strongly induced by treatment with serum after serum starvation in TL-Om1 cells infected with vector control lentivirus, whereas minimal induction was observed in TL-Om1 cells exogenously expressing DUSP5 (Figure 5E). These results suggest that upregulation of DUSP5 suppressed tumor cell growth by inactivating the ERK signaling pathway in ATL cells.
Introduction of DUSP5 expression in ATL cells inhibits cell growth and ERK phosphorylation. (A) Experimental protocol for the establishment of HTLV-1–infected cell lines exogenously expressing DUSP5 and assays. (B) The ratio of GFP-positive cells was monitored at the indicated days, and changes in the GFP population were calculated as follows: 100 × (GFP population at day X)/(GFP population at day 0) based on data from 3 independent experiments. Day 0 was defined as 3 days after lentivirus infection. Gray lines indicate cells infected with empty vector. Blue lines indicate cells infected with lentiviral vectors expressing DUSP5. (C) The total number of GFP-positive cells was calculated from the data of panel B. (D) Immunoblots showing the effect of phosphorylation of ERK1/2 in MT-1 and TL-Om1 cells induced by exogenously expressed DUSP5 under normal growth conditions. Data are expressed as the mean of 3 independent individuals with SD. Differences were tested using the paired t test. (E) Immunoblots showing changes in the phosphorylation pattern of ERK1/2. TL-Om1 cells exogenously expressing DUSP5 were isolated and treated with 10% FBS for the indicated times after serum starvation for 15 hours. FBS, fetal bovine serum; SD, standard deviation.
Introduction of DUSP5 expression in ATL cells inhibits cell growth and ERK phosphorylation. (A) Experimental protocol for the establishment of HTLV-1–infected cell lines exogenously expressing DUSP5 and assays. (B) The ratio of GFP-positive cells was monitored at the indicated days, and changes in the GFP population were calculated as follows: 100 × (GFP population at day X)/(GFP population at day 0) based on data from 3 independent experiments. Day 0 was defined as 3 days after lentivirus infection. Gray lines indicate cells infected with empty vector. Blue lines indicate cells infected with lentiviral vectors expressing DUSP5. (C) The total number of GFP-positive cells was calculated from the data of panel B. (D) Immunoblots showing the effect of phosphorylation of ERK1/2 in MT-1 and TL-Om1 cells induced by exogenously expressed DUSP5 under normal growth conditions. Data are expressed as the mean of 3 independent individuals with SD. Differences were tested using the paired t test. (E) Immunoblots showing changes in the phosphorylation pattern of ERK1/2. TL-Om1 cells exogenously expressing DUSP5 were isolated and treated with 10% FBS for the indicated times after serum starvation for 15 hours. FBS, fetal bovine serum; SD, standard deviation.
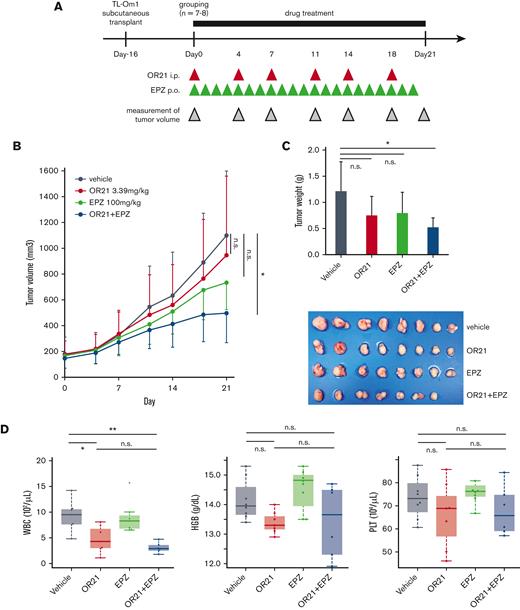
Combination treatment with OR21 and EPZ suppresses tumor cell growth in a xenograft mouse model without inducing myelosuppression
Next, we analyzed the effect of combination treatment with OR21 and EPZ on the growth of TL-Om1 xenograft tumors (Figure 6A). The combination treatment significantly inhibited tumor growth, whereas each single treatment did not have a significant effect (Figure 6B). Mice were sacrificed, and xenograft tumors were isolated 21 days after the initiation of drug treatment. Combination treatment significantly decreased tumor weight (Figure 6C). OR21 induced leukopenia (Figure 6D), and phase 2 studies have shown that EZH2 inhibitors cause lymphopenia and neutropenia.22,42 However, combination treatment did not increase drug-induced hematotoxicity compared with the effect of either single treatment (Figure 6D). We also observed that the combination treatment did not show greater toxicity to neutrophils, monocytes, and lymphocytes than each monotherapy using wild-type mice (supplemental Figure 8). Thus, combination treatment with OR21 and EZH2 inhibitors has a stronger anti-ATL effect than each single treatment without increasing side effects.
Anti-ATL effect of OR21 and EPZ in a TL-Om1 xenograft mouse model. (A) Experimental protocol for the establishment of the TL-Om1 xenograft mouse model and treatment with OR21 and/or EPZ. NOG mice inoculated with TL-Om1 cells subcutaneously were treated with vehicle (1% dimethyl sulfoxide, intraperitoneally, twice per week and 0.5% CMC-Na + 0.1% Tween-80 solution, oral gavage, daily, n = 8), OR21 (3.39 mg/kg, intraperitoneally, twice a week, n = 8), EPZ (100 mg/kg, oral gavage, daily, n = 8), or OR21 + EPZ (3.39 mg/kg OR21 and 100 mg/kg EPZ, n = 7) as indicated by the red and green arrowheads. (B-C) Mean tumor volume (B) and weight (C) in mice. Differences between vehicle and each treatment group were tested using Dunnett test. (D) Concentration of hemoglobin (HGB) and complete blood counts, including white blood cells (WBC) and platelets (PLT), from mice sacrificed at 21 days after the first treatment. Differences between each group were tested using Tukey test. ∗P < .05, ∗∗P < .01. n.s., not significant.
Anti-ATL effect of OR21 and EPZ in a TL-Om1 xenograft mouse model. (A) Experimental protocol for the establishment of the TL-Om1 xenograft mouse model and treatment with OR21 and/or EPZ. NOG mice inoculated with TL-Om1 cells subcutaneously were treated with vehicle (1% dimethyl sulfoxide, intraperitoneally, twice per week and 0.5% CMC-Na + 0.1% Tween-80 solution, oral gavage, daily, n = 8), OR21 (3.39 mg/kg, intraperitoneally, twice a week, n = 8), EPZ (100 mg/kg, oral gavage, daily, n = 8), or OR21 + EPZ (3.39 mg/kg OR21 and 100 mg/kg EPZ, n = 7) as indicated by the red and green arrowheads. (B-C) Mean tumor volume (B) and weight (C) in mice. Differences between vehicle and each treatment group were tested using Dunnett test. (D) Concentration of hemoglobin (HGB) and complete blood counts, including white blood cells (WBC) and platelets (PLT), from mice sacrificed at 21 days after the first treatment. Differences between each group were tested using Tukey test. ∗P < .05, ∗∗P < .01. n.s., not significant.
Discussion
Here, we showed that DNA demethylating agents and EZH2 inhibitors act synergistically to inhibit ATL cell growth. DAC and OR21 were more effective in combination with EPZ than AZA (Figure 1A; supplemental Table 2). AZA exhibits lower DNA demethylating activity than DAC and OR21.24 Particularly in MT-1 cells, the combination of AZA with EPZ did not show a synergistic effect (Figure 1A; supplemental Table 2). In MT-1 cells, AZA is a stronger inhibitor of cell growth than DAC and OR21, whereas its DNA demethylation activity is lower.24 A major fraction of AZA is incorporated into RNA to induce antitumor effects.43,44 The present results suggest that the DNA demethylation activity of these agents contributes to the synergistic effect with EZH2 inhibitors. DS inhibited ATL cell growth at a lower concentration than EPZ when used in combination treatment (Figure 1B). EZH1 and EZH2 are highly expressed in ATL,20 and EZH1 compensates for the decrease in H3K27me3 induced by the inactivation of EZH2.20,21 The present results indicate that dual inhibition of EZH1 and EZH2 is a more effective combination with DNA demethylating agents. In short, both DNA and H3K27 demethylation activities may contribute to the synergistic anti-ATL effect.
As shown in supplemental Figure 2A, half of the genes downregulated in ATL cells with DNA hypermethylation also showed H3K27 hypermethylation. EZH2 recruits DNA methyltransferase, and CpG methylation of EZH2-target promoters is induced.45 This suggests that DNA hypermethylation in ATL may be caused by the recruitment of DNA methyltransferase by EZH2. As shown in Figure 2D-F, genes upregulated by OR21 and EPZ showed both hypermethylated CpG sites and high levels of H3K27me3. The present results indicate that the effect of combination treatment on changing gene expression is mediated by both DNA and H3K27 demethylation activities and not by enhancing each demethylation activity separately (Figure 2A-B,G-H,J). In MT-1 cells, the DNA demethylation activity of combination treatment was slightly lower than that of OR21 (Figures 2A,G-H,J and 4C). The DNA demethylation agent is an S-phase–specific agent.46 Thus, the stronger growth suppression by combination treatment probably contributed to the lower DNA demethylation activity, whereas combination treatment had a greater effect on gene expression than OR21 (Figures 2C-D and 4A). In short, inhibiting both DNA methylation and H3K27me3 accumulation is effective for reprogramming gene expression and thus inhibiting tumor cell growth.
Here, DUSP5 downregulation in ATL cells was caused by promoter CpG methylation and H3K27me3 accumulation (Figure 3A,C,F-H). DUSP5 expression was lower in ATL cell lines than in normal CD4+ T cells (Figure 3D) and was upregulated by the combination of DNA demethylating agents and EZH2 inhibitors (Figure 4A-B). DUSP5 is silenced in cancer cells by the accumulation of H3K27me3 via recruitment of EZH2 to the promoter region.30,47-49 DS decreases H3K27me3 more effectively than EPZ in ATL cell lines,20 and the combination of DAC and DS had a greater effect on inducing DUSP5 expression than EPZ (Figure 4B). Thus, suppressing the histone trimethylation activity of both EZH1 and EZH2 may be important to restore DUSP5 expression in response to combination treatment. DUSP5 is silenced by CpG hypermethylation in CpG islands in gastric cancer,50 whereas CpG methylation in the CpG islands of DUSP5 is not involved in regulating DUSP5 expression in colorectal cancer.51 In ATL cells, the CpG sites at TSS1500-shore, but not TSS200-island, were hypermethylated (Figure 3F-G; supplemental Figure 6A), and OR21 upregulated DUSP5 expression by demethylating the CpG sites at TSS1500-shore but not at TSS200-island (Figure 4; supplemental Figure 6B). Thus, DUSP5 expression may be regulated by CpG methylation of TSS1500-shore in ATL.
DUSP5 expression affects ERK phosphorylation status and cell growth.28,50,52,53 We showed that exogenous DUSP5 expression inhibited ERK phosphorylation and cell growth in ATL cells (Figure 5B-E). The growth inhibitory effect of DUSP5 was stronger in MT-1 cells than in TL-Om1 cells (Figure 5B-C). This could be because of differences in the dependency on ERK signaling for cell growth: p-ERK1/2 levels are higher in MT-1 cells than in TL-Om1 cells54 (supplemental Figure 9). Thus, inactivation of ERK signaling by DUSP5 is involved in the mechanism underlying the effect of combination therapy on inhibiting cell growth. Downregulation of DUSP5 is associated with poor prognosis in several cancers.50,55,56 Here, DUSP5 expression was lower in CADM1+ T cells, which increase during ATL disease progression,24,41 than in CADM1− T cells, regardless of ATL subtype (Figure 3C). Taken together, the results indicate that downregulation of DUSP5 in HTLV-1–infected cells contributes to ATL leukemogenesis and that restoring its expression by combination treatment with DNA demethylating agents and EZH inhibitors has anti-ATL effects.
The combination of DNA demethylating agents and EZH2 inhibitors is effective against several cancers.57,58 This study is the first to demonstrate the effect of this combination in ATL. We showed that combination treatment with OR21 and EZH2 inhibitors inhibited ATL cell growth synergistically and rapidly (Figures 1 and 6B-C). These results indicate that combination therapy may be useful for patients with ATL who are resistant to monotherapy or need immediate therapeutic effects. Although combination therapy is often associated with side effects, we demonstrated that the hematotoxicity of combination treatment with OR21 and EZH2 was not greater than that of single treatment (Figure 6D). OR21 has lower hematotoxicity than DAC,24-26 suggesting that a combination with OR21 rather than DAC could be clinically beneficial.
When used as a monotherapy, the EZH1/2 dual inhibitor DS-3201b (DS, valemetostat) has demonstrated acceptable safety and efficacy in patients with ATL, and a phase 2 clinical trial of DS is ongoing (ClinicalTrials.gov identifier: NCT04102150). We plan to perform a clinical trial of OR21 as a monotherapy for ATL. The present results suggest that dual targeting of aberrant DNA and histone methylation using combination treatment with OR21 and DS is a feasible therapeutic approach for ATL.
Acknowledgments
The authors thank Masao Matsuoka at Kumamoto University and Yasuaki Yamada of Nagasaki University Graduate School of Biomedical Sciences for providing HTLV-1–infected human T-cell lines. Flow cytometric analyses, pyrosequencing, and real-time RT-PCR were conducted at the Analytical Research Center for Experimental Sciences, Saga University.
This work was supported by JSPS KAKENHI grants JP17H06956 and JP20K07593, Yasuda Medical Foundation, and OHARA Pharmaceutical Co., who provided partial financial support.
Authorship
Contribution: Y.K., T.W., and S.K. designed, and Y.K. conducted most experiments; S.Y., N.H. and T.U. performed comprehensive DNA methylation analysis; K.K, Y.F.-K., and Y.Y. contributed to animal experiments and a part of in vitro experiments; T.W., H.N., and E.S. performed some experiments using human clinical samples; T.W., A.K., S.Y., and T.U. performed bioinformatics analyses; T.W. and S.K. obtained funding for this work; and Y.K., T.W., and S.K. wrote the manuscript, with input from all authors.
Conflict-of-interest disclosure: OHARA Pharmaceutical Co. provided partial financial support for this work. Y.K. and Y.F.-K. are full-time employees of OHARA Pharmaceutical Co. T.W. received grants from Nippon Shinyaku. S.K. has received honoraria from Bristol Myers Squibb, Novartis, Pfizer, and Otsuka Pharmaceuticals, and research funding from Bristol Myers Squibb, Novartis, Pfizer, and Otsuka Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Tatsuro Watanabe, Saga University, 5-1-1 Nabeshima, Saga 849-8501, Japan; e-mail: sn6538@cc.saga-u.ac.jp.
References
Author notes
Data are available on request from the corresponding author, Tatsuro Watanabe (sn6538@cc.saga-u.ac.jp).
The full-text version of this article contains a data supplement.