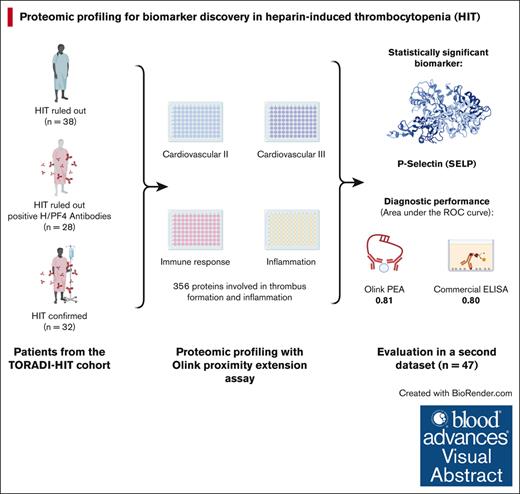
We applied proteomic profiling to patients with suspected HIT, thus analyzing a large number of potential proteins.
Our analysis provided evidence supporting the potential of soluble P-selectin as a promising new biomarker in HIT.
Visual Abstract
New analytical techniques can assess hundreds of proteins simultaneously with high sensitivity, facilitating the observation of their complex interplay and role in disease mechanisms. We hypothesized that proteomic profiling targeting proteins involved in thrombus formation, inflammation, and the immune response would identify potentially new biomarkers for heparin-induced thrombocytopenia (HIT). Four existing panels of the Olink proximity extension assay covering 356 proteins involved in thrombus formation, inflammation, and immune response were applied to randomly selected patients with suspected HIT (confirmed HIT, n = 32; HIT ruled out, n = 38; and positive heparin/platelet factor 4 [H/PF4] antibodies, n = 28). The relative difference in protein concentration was analyzed using a linear regression model adjusted for sex and age. To confirm the test results, soluble P-selectin was determined using enzyme-linked immunosorbent assay (ELISA) in above mentioned patients and an additional second data set (n = 49). HIT was defined as a positive heparin-induced platelet activation assay (washed platelet assay). Among 98 patients of the primary data set, the median 4Ts score was 5 in patients with HIT, 4 in patients with positive H/PF4 antibodies, and 3 in patients without HIT. The median optical density of a polyspecific H/PF4 ELISA were 3.0, 0.9, and 0.3. Soluble P-selectin remained statistically significant after multiple test adjustments. The area under the receiver operating characteristic curve was 0.81 for Olink and 0.8 for ELISA. Future studies shall assess the diagnostic and prognostic value of soluble P-selectin in the management of HIT.
Introduction
Diagnostic workup, assessment of prognosis, and treatment monitoring of heparin-induced thrombocytopenia (HIT) are hampered by a lack of reliable and specific biomarkers. HIT is a severe adverse reaction to heparin, one of the most commonly used anticoagulants.1 Exposure to heparin can trigger the formation of platelet-activating antibodies against a heparin-platelet factor 4 (PF4) complex.2-5 Paradoxically, these antibodies can induce a prothrombotic state, leading to severe thromboembolism, limb loss, and even death.6 In contrast, patients suspected of having HIT are often treated with dangerous anticoagulants with a high bleeding risk, such as argatroban.7-9 Thus, misdiagnosis of HIT has severe consequences, including increased morbidity and mortality due to overtreatement or undertreatment.10 Due to their limited availability and prolonged turnaround times, washed platelet activation assays, which are regarded as the reference standard, are not suitable for use in the acute phase of HIT.11,12 The commonly used heparin/PF4 (H/PF4) antibody assays, however, have limited specificity and, therefore, put the patient at risk of overtreatment.13 Despite recent advancements, including automated H/PF4 antibody assays, prediction models, and machine-learning applications, there is still a diagnostic gap that needs to be addressed.14-17 Therefore, new biomarkers are a promising tool to develop enhanced diagnostic tests for the diagnosis, prognosis, or monitoring of HIT.18
New analytical techniques enable the simultaneous determination of hundreds of biomarkers with extremely high sensitivity.19 Proteins are critical mediators in hemostasis mechanisms, contributing to immunological response and inflammation, and venous and arterial thromboembolism.20 These techniques can help in observing the interplay of protein-biomarkers and their role in the mechanism of HIT. Among these techniques, Olink proximity extension assay (PEA; Uppsala, Sweden) for proteomic profiling stands out for its high sensitivity, low risk of interferences, low specimen volume, and the large number of biomarkers that can be determined simultaneously.21 This powerful platform has already been used successfully to identify potential biomarkers for a range of diseases, including cardiovascular disease, inflammatory diseases, cancer, and infectious diseases.22-25
We hypothesize that the application of proteomic profiling using the Olink PEA platform can identify novel biomarkers for the management of HIT, potentially enabling a more accurate diagnosis.
Methods
Study design, setting, and population
This analysis was conducted in line with a large prospective cohort study. Three groups of patients were selected out of 120 patients recruited in line with the TORADI-HIT data set16,26 or a preceding pilot study:11,27 (1) confirmed HIT; (2) H/PF4 antibodies present but HIT ruled out; and (3) HIT ruled out, H/PF4 antibodies not present (Figure 1; primary data set). Patients in each group were randomly selected. An additional, random sample of 50 patients was selected to confirm the findings in a second data set. Overall inclusion criteria were as follows: (1) suspected HIT, which included anti-H/PF4 antibody assay ordered or 4Ts score rated or hematology consultancy service requested; (2) age ≥18 years; and (3) general informed consent. Exclusion criteria were (1) insufficient serum specimen, (2) insufficient clinical data, and (3) did not pass Olink quality control. The TORADI-HIT study recruited patients from 11 study centers in Switzerland, Germany, and the United States.16 Most patients were included in Inselspital, University Hospital of Bern, Bern, Switzerland. Biomarker discovery was done using Olink PEA (356 different proteins). The results were verified using enzyme-linked immunosorbent assay (ELISA) determinations of the proteins (at French Blood Establishment [EFS] Auvergne-Rhone-Alpes, and University Jean Monnet, Mines Saint-Etienne, and INSERM, U 1059 SAINBIOSE laboratory).
Definition of patient groups
HIT was defined by a positive washed platelet functional assay, specifically the heparin-induced platelet activation assay (HIPA).11,16,27 Multiple studies have demonstrated that washed platelet assays, such as the serotonin release assay and HIPA, exhibit high sensitivity and specificity, and strong concordance with clinical HIT. Therefore, the American Society of Hematology and the British Committee for Standards in Hematology recommend these assays as reference standards.28,29 Patients with positive H/PF4 antibodies were defined by a positive immunoassay (ELISA) but a negative HIPA. Patients with negative HIT were defined by a negative ELISA and a negative HIPA.
Workup and laboratory tests
Detailed clinical and laboratory data including residual serum samples were collected at diagnosis following a prespecified protocol. Serum samples were frozen at –80°C. HIPA and H/PF4 immunoassays were conducted within 1 week after arrival. The laboratory technicians were blinded to the results of the other test and to the clinical information.
For the HIPA, serum samples were incubated with 4 different washed platelet donations in the presence of (1) only buffer, (2) 0.2 IU/mL low molecular weight heparin, and (3) 100 IU/mL heparin. All details were published previously.11,16,30 The test was considered positive if aggregation occurred within 30 minutes for at least 2 donors in the presence of 0.2 IU/mL low molecular weight heparin but not in the presence of 100 IU/mL heparin. On each plate, positive and negative controls were also measured.
For the H/PF4 immunoassay, the polyspecific Lifecodes PF4 Enhanced (Immucor, Dreieich, Germany) was performed according to the manufacturer’s instructions. Optical density >0.5 was considered positive. The test was previously validated in our laboratory, and external and internal quality controls were performed.11
Proteomic profiling
To assess the proteomic profile, 4 existing panels of Olink (Olink Proteomics Inc, Uppsala, Sweden) PEA were performed by Olink Uppsala: “Cardiovascular II,” “Cardiovascular III,” “Immune response,” and “Inflammation.” These panels include 356 different proteins involved in thrombus formation and inflammation. A full list of all proteins can be found in supplemental Table 3. In short, the PEA recognizes proteins by pairs of oligonucleotide-linked antibodies.21 If the antibodies bind in proximity to each other, the oligonucleotides hybridize, and a new polymerase chain reaction primer sequence is revealed. This DNA barcode is then amplified and detected via quantitative polymerase chain reaction. The cycle threshold value, which is inversely correlated to the protein concentration in the sample, is then normalized and transformed to an arbitrary unit called normalized protein expression on a log 2 scale. The quality of the measurements is assured through multiple internal controls (incubation controls, extension controls, and detection controls) as well as sample controls (interplate and negative controls), details of which are described elsewhere.31 This innovative technique has been successfully used to identify various key biomarkers in a broad range of diseases, including venous thromboembolism.22-25,32 The proteins were then annotated with their corresponding gene using the Human Protein Atlas project.33
P-selectin ELISA technology
The levels of soluble P-selectin (soluble CD62P; corresponding to SELP; minimum detectable concentration, 0.244 ng/mL) were quantified in serum samples using ELISA (IBL International, Hamburg, Germany). Absorbance at 450 nm (for serotonin, 405 nm) was measured using an ELISA plate reader (Magellan Software; SUNRISE TIME, Tecan Group Ltd, Lyon, France). Results were normalized to 2 × 108 platelets per mL, and data were expressed in pg/mL.34
Statistical analysis
To explore the variability between different patient groups, a principal component analysis (PCA) using single value decomposition and sparse least square analysis (sPLS) was used. Additionally, to quantify the association between protein levels and the presence of HIT, we fitted a linear model to the data using the “stats” package for R. In the model, the normalized protein expression value of the different proteins served as the dependent variable, whereas the HIPA status was used as the independent variable. To account for physiological differences among patients, the model was adjusted for age and sex. The Benjamini-Hochberg method was used to adjust the calculated P value to account for multiple testing, setting the false discovery rate at 5%. A heat map showing the 50 most significantly changed proteins and a volcano plot were plotted. For the biomarker that showed the highest significance, we created box plots by thrombosis status and compared the different groups using the Wilcoxon rank-sum test. Finally, to determine the diagnostic usefulness of the biomarker, we performed a receiver-operator characteristic curve (ROC) analysis and calculated the area under the curve (ROC-AUC). Additionally, we performed a multivariable linear regression and ROC analysis using thrombosis as the dependent variable. All analyses were done in R version 4.1.2.
The appropriate ethical committee approved the final protocol (Kantonale Ethikkommission Bern). The study was conducted in accordance with the Declaration of Helsinki.
Results
Patient characteristics
Of a random sample of 120 patients, 32 with confirmed HIT, 28 with H/PF4 detected (without HIT), and 38 without HIT were included (Figure 1; primary data set). Overall, 21 were excluded because of insufficient clinical data or leftover sample material; 1 sample did not pass Olink quality control. The median 4Ts score was 5 in patients with HIT (interquartile range [IQR], 4-6), 4 in patients with positive H/PF4 antibodies (IQR, 3.75-4), and 3 in patients without HIT (IQR, 2-4). The median H/PF4 ELISA was 3.0 (IQR, 2.4-3.0) in patients with HIT, 0.9 (IQR, 0.7-1.5) in patients with positive H/PF4 antibodies, and 0.3 (IQR, 0.2-0.3) in patients without HIT. Detailed patient characteristics are given in Table 1. From the second data set comprising 50 patients with suspected HIT, 1 was excluded because of insufficient data (Figure 1). Among these patients, 12 were HIT positive, 16 were H/PF4 positive, and 21 were HIT negative. Detailed data of this second data set are available in supplemental Table 1.
Patient characteristics of the primary data set
| . | HIT positive . | H/PF4 positive . | HIT negative . |
|---|---|---|---|
| Number, n | 32 | 28 | 38 |
| Male sex, n (%) | 22 (68.8) | 17 (60.7) | 24 (63.2) |
| Age, median (IQR) | 68.5 (64.8-76.0) | 77.0 (55.0-79.0) | 74.0 (54.0-81.0) |
| 4Ts, median (IQR) | 5 (4-6) | 4 (4-5) | 3 (2-4) |
| ELISA GTI polyspecific OD, median (IQR) | 3.0 (2.4-3.0) | 0.9 (0.7-1.5) | 0.3 (0.2-0.3) |
| Setting, n (%) | |||
| Cardiac surgery | 13 (40.6) | 3 (10.7) | 4 (10.5) |
| ICU | 10 (31.2) | 12 (42.9) | 14 (36.8) |
| Others | 9 (28.1) | 13 (46.4) | 20 (52.6) |
| Thrombocytes, median (IQR), x 109/L | 60 (43-81) | 68 (48-101) | 59 (41-80) |
| Thrombosis, n (%) | 15 (57.7) | 5 (25.0) | 4 (11.8) |
| . | HIT positive . | H/PF4 positive . | HIT negative . |
|---|---|---|---|
| Number, n | 32 | 28 | 38 |
| Male sex, n (%) | 22 (68.8) | 17 (60.7) | 24 (63.2) |
| Age, median (IQR) | 68.5 (64.8-76.0) | 77.0 (55.0-79.0) | 74.0 (54.0-81.0) |
| 4Ts, median (IQR) | 5 (4-6) | 4 (4-5) | 3 (2-4) |
| ELISA GTI polyspecific OD, median (IQR) | 3.0 (2.4-3.0) | 0.9 (0.7-1.5) | 0.3 (0.2-0.3) |
| Setting, n (%) | |||
| Cardiac surgery | 13 (40.6) | 3 (10.7) | 4 (10.5) |
| ICU | 10 (31.2) | 12 (42.9) | 14 (36.8) |
| Others | 9 (28.1) | 13 (46.4) | 20 (52.6) |
| Thrombocytes, median (IQR), x 109/L | 60 (43-81) | 68 (48-101) | 59 (41-80) |
| Thrombosis, n (%) | 15 (57.7) | 5 (25.0) | 4 (11.8) |
ICU, intensive care unit; OD, optical density.
Proteomic profile
The primary data set was used for proteomic profiling. In PCA and sPLS, minor differences between HIPA-positive and HIPA-negative patients were observed. Overlapping clusters were interpreted as a consequence of low patient numbers and similar patient characteristics (patients with suspected HIT). Results of the PCA and sPLS are displayed in supplemental Figures 1 and 2.
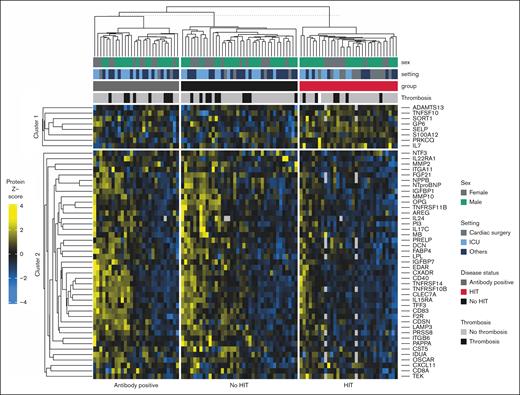
Protein abundance analysis revealed a statistically significant association of 40 proteins with HIT status (8 upregulated and 32 downregulated). Of these proteins, soluble P-selectin remained statistically significant after multiple test adjustments (false discovery rate, 5%; = 1.04; 95% confidence interval [CI], 0.63-1.45). A clustered heat map is available in Figure 2, and a volcano plot showing adjusted P value is available in Figure 3. Fold changes with adjusted P value are available in the supplemental Material.
Clustered heat map illustrating the z scores of the 50 most significant proteins, stratified by HIT status (group). The following additional information is shown: sex, setting, and presence of thrombosis. ICU, intensive care unit.
Clustered heat map illustrating the z scores of the 50 most significant proteins, stratified by HIT status (group). The following additional information is shown: sex, setting, and presence of thrombosis. ICU, intensive care unit.
Volcano plot showing the differential abundance of proteins between patients with HIT and those without HIT (including H/PF4 antibody positives). The x-axis depicts the fold change (normalized protein expression [NPX] difference), whereas the y-axis depicts the –log10(adjusted P value). Green dots represent a P value < .05, and red dots represent adjusted P value between .05 and 0.3.
Volcano plot showing the differential abundance of proteins between patients with HIT and those without HIT (including H/PF4 antibody positives). The x-axis depicts the fold change (normalized protein expression [NPX] difference), whereas the y-axis depicts the –log10(adjusted P value). Green dots represent a P value < .05, and red dots represent adjusted P value between .05 and 0.3.
ELISA and additional analyses
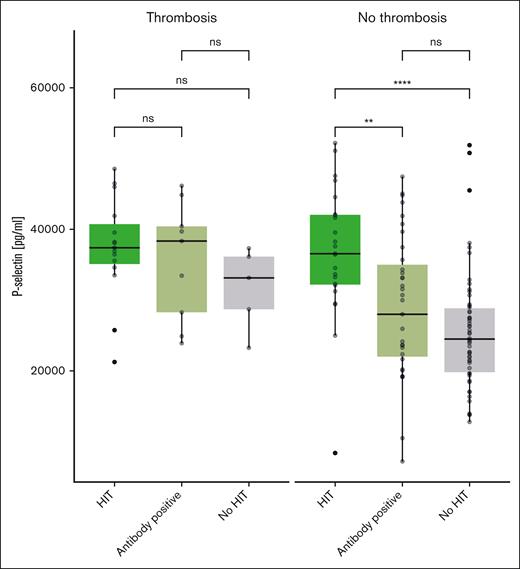
An ELISA was used to determine the serum soluble P-selectin levels both in the primary data set that underwent Olink PEA and in an additional data set of 49 patients suspected of having HIT. First, we analyzed the first data set and found the following median soluble P-selectin values: 25 783 pg/mL (IQR, 21 238-27 157) for patients without HIT, 29 350 pg/mL (IQR, 22 175-36 963) for patients with negative HIPA but positive immunoassay, and 38 150 pg/mL (IQR, 33 888-42 075) for patients with HIT. There was a statistically significant difference between all groups when compared with the patients without HIT (no HIT vs antibody positive, P = .02; no HIT vs HIT, P ≤ .01).
Interestingly, different results are seen when analyzing only patients with thromboembolism: 32 423 pg/mL (IQR, 27 342-36 437), 33 450 pg/mL (IQR, 24 900-34 650; P = .73), and 37 750 pg/mL (IQR, 35 988-42 925; P = .13) for patients with negative HIT, positive antibody, and positive HIT, respectively.
Similar results were obtained in the second, confirmatory data set: 24 147 pg/mL (IQR, 19 627-24 149) in patients without HIT, 31 547 pg/mL (IQR, 24 057-31 342; P = .02) in patients with positive antibodies, and 35 048 pg/mL (IQR, 32 038-38 087; P ≤ .01) in patients with HIT. In contrast, no significant differences were seen in patients with thromboembolism. Box plots showing soluble P-selectin levels for both data sets combined are displayed in Figure 4.
P-selectin in patients with HIT, positive H/PF4 antibodies, and without HIT, depending on the presence of thromboembolism (ELISA, all patients).
P-selectin in patients with HIT, positive H/PF4 antibodies, and without HIT, depending on the presence of thromboembolism (ELISA, all patients).
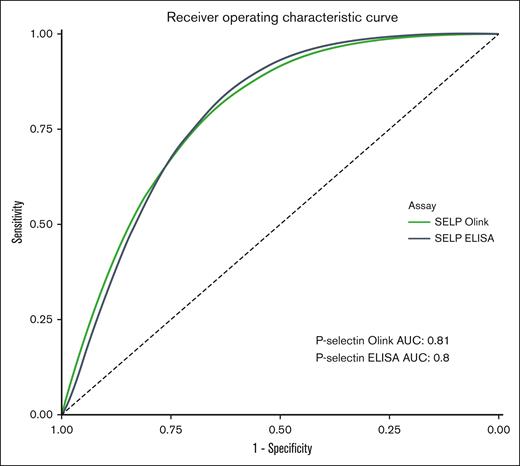
ROC analysis of the soluble P-selectin as measured by Olink PEA in the first cohort showed a ROC-AUC of 0.81 (95% CI, 0.72-0.90) Similar results were observed with the ELISA (ROC-AUC, 0.80; both groups). The results are illustrated in Figure 5.
ROC curve of P-selectin for the presence of HIT as measured with the Olink PEA and ELISA (all patients).
ROC curve of P-selectin for the presence of HIT as measured with the Olink PEA and ELISA (all patients).
ROC analysis of soluble P-selectin for detecting thrombosis showed a lower ROC-AUC of 0.65 (95% CI, 0.52-0.77) for the Olink PEA and 0.67 (95% CI, 0.55-0.79) for the ELISA (supplemental Figure 3). An additional multivariable linear regression showed a significant association between P-selectin levels and the different patient groups, even when adjusting for the presence of thrombosis, age, and sex (supplemental Table 2).
Discussion
We applied the Olink PEA, covering 356 proteins involved in thrombus formation, inflammation, and immune response, to 98 randomly selected patients with suspected HIT and confirmed the results with ELISA in the patients mentioned above and an additional data set of 47 patients. Among 40 proteins that were statistically significantly associated with HIT status in protein abundance analysis, soluble P-selectin remained significant after multiple test adjustments. This association was confirmed in a ROC analysis in PEA and ELISA (0.80 and 0.81, respectively). This association was especially apparent in patients without thrombosis, suggesting potential usefulness in this group.
To our knowledge, this is the first investigation to apply the PEA technology to patients with suspected HIT, thus analyzing a large number of proteins potentially associated with immune-mediated thrombosis. Prior omics-based analyses primarily focused on genetic variants. Four genome-wide association studies investigated the risk factors of HIT and revealed genetic variants associated with various enzymes, the AB0 complex, and distinct receptor proteins.18,35-37 However, comprehensive studies including metabolomics, proteomics, and transcriptomics are still missing.18
Our study suggests that soluble P-selectin holds potential as a diagnostic marker for HIT. P-selectin is a glycoprotein that is expressed in platelets and endothelial cells and is involved in leukocyte adhesion and thrombocyte aggregation.38 When platelets are activated, P-selectin is mobilized from the α-granules to the external membrane.39 In recent years, this mechanism has been leveraged to develop flow cytometry–based tests for activated platelets in patients suspected of having HIT. However, the diagnostic performance of these tests is limited.27,40 Besides, soluble P-selectin, which can be released into the bloodstream through proteolytic cleavage or alternative splicing, has been shown to be elevated in various cardiovascular and thrombotic disorders, including myocardial infarction, venous thrombosis, and COVID-19–related thrombosis.41-45 Thus, soluble P-selectin appears to be a general marker for platelet activation.46 Moreover, CD62P-mediated cross talk between the vessel wall, platelets, monocytes, and neutrophils results in the activation of innate immune cells and an increase in the expression of tissue factor. This initial activation of immune cells has the effect of thrombus reinforcement and retardation of subsequent resolution processes.32 Interestingly, our findings extend decades-old observations on increased values of soluble P-selectin in patients with HIT.47-49 However, these studies have methodological limitations, and soluble P-selectin was not yet considered a biomarker for HIT.
Our study has several strengths. Most importantly, the patients were randomly selected from a population of patients with suspected HIT. This is closely resembling the target population for a potential diagnostic or prognostic test, including not only patients with confirmed HIT but also patients with H/PF4 antibodies and patients without HIT but with similar presenting diseases. As a consequence, contrasts are less pronounced compared with healthy controls but correspond to realistic clinical settings. In addition, we analyzed a large number of proteins in a relatively large cohort. Besides, the results obtained with the PEA were confirmed with an independent analytical technique (ELISA) and in a second data set. All of these points contribute to the high validity of the study.
However, our study also has some limitations. Firstly, we excluded a certain proportion of patients due to incomplete clinical data or missing sample material. However, we consider these dropouts to be at least "at random" and thus unlikely to affect the results of the study. Secondly, the population was not consecutive because of the high costs of the PEA tests. Thus, we cannot fully exclude that a certain selection bias is present. One might additionally argue that a matching procedure according to age and sex would increase the validity of the results. To account for this, we included age and sex in the regression model. These limitations suggest that our results must be confirmed in an independent, larger cohort of consecutive patients. Such a diagnostic accuracy study would also have to be carried out with test systems that can be used in daily practice (eg, chemiluminescent immunoassay). Another limitation is that we have used serum rather than plasma. However, this limitation is minimized due to the differential analysis of the various groups using the same sample preparation process. In addition, a polyspecific rather than a immunoglobulin-specific ELISA was used. However, there are 3 reasons why this point did not introduce bias: (1) it is not a diagnostic accuracy study in which the performance of current tests is used as a comparison; (2) several studies have shown that the correlation between polyspecific and immunoglobulin-specific immunoassays is very high;13 and (3) this information is not included in the study either as an investigated variable or as an outcome variable.
Our data confirm that soluble P-selectin is a promising new biomarker in patients with HIT. This fits with our current understanding of the mechanism of HIT, which recognizes platelet activation as an important feature. The concept of soluble P-selectin as a general biomarker for platelet activation is supported by comparable observations in many other thromboembolic diseases. Soluble P-selectin might be included in future diagnostic decision-support tools, thus adding information about platelet activation. The protein may be particularly useful in the diagnosis of HIT without thrombosis, which is a particularly challenging diagnostic situation. Furthermore, the differential concentration of soluble P-selectin in patients with and without thrombosis suggests its potential use as a prognostic marker. This is consistent with observations suggesting soluble P-selectin as a prognostic marker for thromboembolism in COVID-19.45 However, our findings must be confirmed in future studies, prospectively including patients with suspected HIT.
In conclusion, our analysis of 356 proteins associated with thrombus formation, inflammation, and immune response in a representative study of patients with suspected HIT has provided evidence supporting the potential of soluble P-selectin as a promising new biomarker. Because this was particularly apparent in patients without thrombosis, a potential application appears not only as a diagnostic but also as a prognostic biomarker. Nevertheless, further validation of our findings in diverse settings and populations is warranted, necessitating prospective studies that include patients with suspected HIT.
Acknowledgments
The authors thank Justine Brodard for implementing the heparin-induced platelet activation at Inselspital, Vincent Benites and Laura Celeste Rotondo for performing all laboratory tests, Anja Stalder and Margret Bachmann-MacDonald for study management, and residents at all study centers. The authors acknowledge those individuals who provided technical support throughout the investigations including Charles-Antoine Arthaud, Marie-Ange Eyraud, and Amelie Prier from the Etablissement Français du Sang (EFS) Auvergne-Rhone-Alpes, Saint-Etienne, France.
This study was supported by a research grant from the Swiss National Science Foundation (215574), the International Society on Thrombosis and Haemostasis, and the Clinical Trial Unit research grants from Inselspital, University Hospital.
Authorship
Contribution: H.N. wrote the analysis plan, analyzed and interpreted the data, and wrote the first draft of the manuscript; H.H.-C. and F.C. contributed to the design of the study, analyzed and interpreted data, provided infrastructure and reagents, and contributed to the first draft of the manuscript; J.H. contributed to the analysis plan and interpretation of data; J.-D.S., A.G., D.A.T., A.M., W.A.W., A.S., J.A.K.H., B.G., P.V., T.B., and L.G. collected data; M.N. designed and implemented the study, collected data, contributed to analysis plan and interpretation of data, and wrote the manuscript; and all authors contributed to the interpretation of data, reviewed the manuscript critically, and approved the final version of the manuscript.
Conflict-of-interest disclosure: The institution of J.A.K.H. received grant support, consultancy fees, or honoraria from Swiss National Science Foundation, Baxter/Takeda, Bayer, CSL Behring, Novo Nordisk, Octapharma, Roche, Sobi, Roche, Sanofi, Federal Office of Public Health, and Swiss Hemophilia Society, outside of the present work. M.N. received research grants from Bayer Healthcare, Roche Diagnostics, Siemens Healthineers, Pentapharm, and BÜHLMANN Laboratories, as well as lecture fees from Sysmex, Siemens Healthineers, and Euroimmun, outside of the present work. A.G. reports personal fees from Aspen, Bayer Vital, CHROMATEC, Instrumentation Laboratory, Macopharma, Sanofi-Aventis, Roche, Gesellschaft für Thrombose- und Hämostaseforschung, Dilaflor, Takeda Pharma, Falk Foundation e.V., and Mylan Germany; grants from Ergomed, Boehringer Ingelheim, ROVI, Sagent, Macopharma, Portola, Biokit, Blau Farmaceutica, Prosensa/BioMarin, Deutsches Rotes Kreuz-Blutspendedienst Baden-Würtemberg/Hessen, Deutsche Forschungsgemeinschaft, Robert-Koch-Institut, Deutsche Gesellschaft für Internationale Zusammenarbeit Else-Körner-Stiftung, and European Medicines Agency; grants and other support from Deutsches Rotes Kreuz-Blutspendedienst der Landesverbände des Deutschen Roten Kreuz Niedersachsen, Sachsen-Anhalt, Thüringen, Oldenburg und Bremen; nonfinancial support from Veralox, Vakzine Projekt Management GmbH, AstraZeneca, and Janssen Vaccines & Prevention B.V., outside the submitted work; and has a patent "Screening methods for transfusion related acute lung injury (TRALI),” with royalties paid to EP2321644, 8 May 2011, and a patent “Verfahren und Vorrichtung zur Herstellung von Universalplasma,” licensed to DE 10 2020 212 609 B3, 7 April 2022. T.B. reports grant support, consultancy fees, honoraria, or support for attending meetings from Deutsche Forschungsgesellschaft, Stiftung Transfusionsmedizin und Immunhämatologie e.V, DRK Blutspendedienst, Deutsche Herzstiftung, Ministerium für Wissenschaft, Forschung und Kunst Baden-Würtemberg, Gesellschaft für Thrombose- und Hämostaseforschung, Berufsverband Deutscher Internisten, CoaChrom Diagnostica GmbH, Robert Bosch GmbH, Ergomed, Bayer, Bristol Myers Squibb, Doctrina Med AG, Leo Pharma GmbH, Schöchl medical education GmbH, Mitsubishi Tanabe GmbH, Novo Nordisk GmbH, and Swedish Orphan Biovitrium GmbH. The remaining authors declare no competing financial interests.
Correspondence: Michael Nagler, Department of Clinical Chemistry, Inselspital, Bern University Hospital, and University of Bern, Freiburgerstrasse 10, 3010 Bern, Switzerland; email: michael.nagler@insel.ch.
References
Author notes
H.N. and H.H.-C. are joint first authors.
Data are available on request from the corresponding author, Michael Nagler (michael.nagler@insel.ch).
The full-text version of this article contains a data supplement.


![Volcano plot showing the differential abundance of proteins between patients with HIT and those without HIT (including H/PF4 antibody positives). The x-axis depicts the fold change (normalized protein expression [NPX] difference), whereas the y-axis depicts the –log10(adjusted P value). Green dots represent a P value < .05, and red dots represent adjusted P value between .05 and 0.3.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/11/10.1182_bloodadvances.2024012782/2/m_blooda_adv-2024-012782-gr3.jpeg?Expires=1769115544&Signature=D0Ql0w7GrbGhSyL6i07q~CZuT59lesTOFtwweugVISc39CSRgn7G9Dq9TuM-Kax0T~QHsJxl6Q3dh74ZXuCwag94ojmZ~2bdka43h7zsPwZhnUW0rMGLabYmoWxAfbFCjlhJdd3XKo7vKjkqS1H7nQsD2~PtiVr7BNC4W8wyPYfQ7cmjDwYJo02AuKhucB9hrL84E~CNnC0ElLc5JXslt8o6sTxOFNvli04l97RZC0cIsVUt9tuMT2dHZ6WGE878ThbDk5JIFF-35NHV3nbxBX79XWLa8cboJ7IC3s4t7abgh1mmAuISShir8ET5NRxjg72swm0OaaEj4oBY1obvQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)