Visual Abstract
TO THE EDITOR:
Hyper(re)active platelets contribute to the development of cardiovascular disease associated with chronic inflammatory and metabolic disorders.1,2 The weak platelet agonists adenosine 5’-disphosphate (ADP) and epinephrine (epi) share common Gi/z protein–coupled receptor signaling pathways via P2Y12 and the α2A-adrenergic receptor, respectively. This includes the inhibition of the adenylate cyclase by the dissociated Gi-protein α subunit, thereby counteracting the formation of the inhibitory secondary messenger cyclic adenosine-monophosphate.3,4 Platelet aggregation in response to low doses of ADP plus arachidonic acid is associated with a higher outcome risk of myocardial infarction and stroke.5 Mammen and Bick developed laboratory criteria for patients with unexplained arterial and venous thrombotic events and platelet hyperaggregability in response to low concentrations of ADP and epi, classified as heritable sticky platelet syndrome.6 However, conclusive data and robust clinical studies to establish this syndrome as a distinct hemostatic disorder, are still missing. Pathologically intrinsic platelet high responsiveness (HR) can cause high on-treatment platelet reactivity to P2Y12 antagonists.7 Based on the common Gi-protein signaling of the P2Y12 and α2A-adrenergic receptors, high reactivity to epinephrine may cause a bypass effect to P2Y12-inhibitor therapy.8
This association study, including single-day blood samples, aims to compare platelet ADP and epi HR in light transmission aggregometry (LTA) with other platelet activation/function tests regulated by Gi/z protein–coupled receptor signaling and to determine the predictive value of clinical factors and platelet-related common genetic variants.
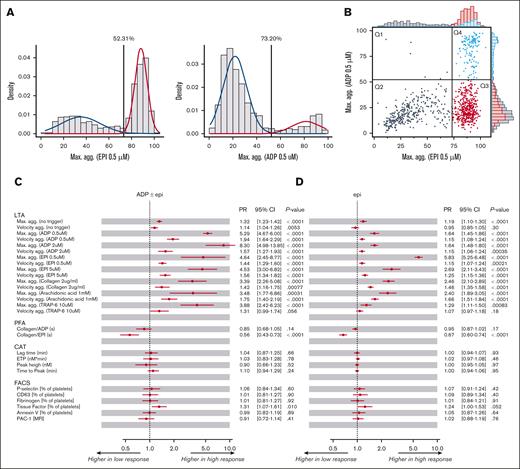
A subcohort of 903 participants with clinical, platelet function, and genetic data from a representative Western European population, the Gutenberg Health Study, were included in this explorative study (supplemental Figure 1). Method details are described in supplemental Material. The baseline characteristics revealed a sex-matched cohort with an average age of 61.8 ± 11.4 years and a prevalence of arterial hypertension, followed by dyslipidemia, obesity, cancer, and diabetes. Prevalent medications, also known to influence platelet function, include cardiovascular system drugs (C), lipid-modifying agents (C10), antiplatelet agents (B01AC), anti-inflammatory/antirheumatic products (M01A), and diabetes drugs (A10) (supplemental Table 1). Low- vs high-responsive platelets to 0.5 μM of ADP and epi, established with low platelet response in the LTA, were discriminated by the intersection of 2 distributions in a Gaussian mixture model, resulting in a cutoff value of 52.31% and 73.20% maximum aggregation, respectively (Figure 1A). Only 0.6% of the study participants (6/903; Figure 1B, Q1) exhibit ADP and not epi high-responsive platelets, whereas 47.1% (425/903; Figure 1B, Q3) showed only epi high-responsive platelets, and 16% (140/903; Figure 1B, Q4) displayed hyperaggregability for both ADP and epi, indicating that nearly all participants with platelet ADP hyperreactivity are also high responsive to epi.
Platelet high reactivity in the LTA induced by a low-response concentration (0.5 μM) of ADP and epi and association with other platelet function parameters. (A) Discrimination of high and low responders to platelet aggregation induced by ADP and epi. The cutoffs for defining platelets from participants as low-responsive and high-responsive to low-response concentrations of ADP (0.5 μM) and epi (0.5 μM) measured by LTA were established as an intersection of 2 Gaussian distributions. Left: blue curve: mean = 21.72, SD = 10.06, weighting in the Gaussian mixture distribution = 0.84. Red curve: mean = 81.65, SD = 12.38, weighting in the Gaussian mixture distribution = 0.16, cutoff = 52.31%. Right: blue curve: mean = 35.22, SD = 17.24, weighting in the Gaussian mixture distribution = 0.38. Red curve: mean = 88.35, SD = 5.53, weighting in the Gaussian mixture distribution = 0.62, cutoff = 73.2%. (B) Distribution of study participants for maximal platelet aggregation responses to low-response concentrations of ADP (0.5 μM) and epi (0.5 μM). Four groups of study participants with distinct platelet reactivity patterns to low concentrations of ADP and epi were identified (Q1-Q4): Q1, high responsive to ADP and low responsive to epi; Q2, low responsive to both ADP and epi; Q3, low responsive to ADP and high responsive to epi; Q4, high responsive to both ADP and epi. (C-D) Associations between platelet aggregation in response to low-response concentrations of ADP (0.5 μM) and/or epi (0.5 μM) and parameters of other platelet function tests regulated by Gi protein–coupled receptor signaling. (C) ADP epi, (D) epi were investigated using Poisson regressions with robust standard errors. The model was adjusted for age, sex, platelet count (whole blood), and platelet function-interfering medications. CAT, calibrated automated thrombinography; CI, confidence interval; FACS, flow cytometry; MFI, mean fluorescence intensity; PFA, platelet function analyzer; TRAP-6, thrombin receptor–activated peptide 6.
Platelet high reactivity in the LTA induced by a low-response concentration (0.5 μM) of ADP and epi and association with other platelet function parameters. (A) Discrimination of high and low responders to platelet aggregation induced by ADP and epi. The cutoffs for defining platelets from participants as low-responsive and high-responsive to low-response concentrations of ADP (0.5 μM) and epi (0.5 μM) measured by LTA were established as an intersection of 2 Gaussian distributions. Left: blue curve: mean = 21.72, SD = 10.06, weighting in the Gaussian mixture distribution = 0.84. Red curve: mean = 81.65, SD = 12.38, weighting in the Gaussian mixture distribution = 0.16, cutoff = 52.31%. Right: blue curve: mean = 35.22, SD = 17.24, weighting in the Gaussian mixture distribution = 0.38. Red curve: mean = 88.35, SD = 5.53, weighting in the Gaussian mixture distribution = 0.62, cutoff = 73.2%. (B) Distribution of study participants for maximal platelet aggregation responses to low-response concentrations of ADP (0.5 μM) and epi (0.5 μM). Four groups of study participants with distinct platelet reactivity patterns to low concentrations of ADP and epi were identified (Q1-Q4): Q1, high responsive to ADP and low responsive to epi; Q2, low responsive to both ADP and epi; Q3, low responsive to ADP and high responsive to epi; Q4, high responsive to both ADP and epi. (C-D) Associations between platelet aggregation in response to low-response concentrations of ADP (0.5 μM) and/or epi (0.5 μM) and parameters of other platelet function tests regulated by Gi protein–coupled receptor signaling. (C) ADP epi, (D) epi were investigated using Poisson regressions with robust standard errors. The model was adjusted for age, sex, platelet count (whole blood), and platelet function-interfering medications. CAT, calibrated automated thrombinography; CI, confidence interval; FACS, flow cytometry; MFI, mean fluorescence intensity; PFA, platelet function analyzer; TRAP-6, thrombin receptor–activated peptide 6.
Four group comparisons were performed with participants rather reflecting high-responsive platelets to ADP (ADP ± epi HR vs normoreactive [ADP ± epi]; epi ± ADP HR vs epi HR [epi ± ADP vs epi]) and epi (epi ± ADP HR vs normoreactive [epi ± ADP]; epi HR vs normoreactive [epi]), respectively. Using robust Poisson regression analysis, estimated prevalence ratios (PRs) revealed that increased platelet aggregability in response to ADP and/or epi was associated with an increased maximum of aggregation for all tested agonists at high signaling concentrations (Prange, .019 to <.0001) and percentage of tissue factor-positive platelets in vivo (Prange, .083-.0033), adjusted for age, sex, platelet count (weak correlation between platelet ADP/epi–induced aggregation and platelet count; supplemental Figure 2), and platelet function-interfering medications (Figure 1C-D; supplemental Figure 3). PRs for collagen-epinephrine (CEPI) closure time in the platelet function analyzer decreased in participants with platelet hyperaggregability. Notably, treatment with antiplatelet agents was prominent in participants with low-responsive platelets to 0.5 μM ADP/epi in the LTA, associated with prolonged CEPI closure times, whereas reference CEPI closure times were observed for high-responsive platelets to 0.5 μM ADP/epi in the LTA in participants with sporadic antiplatelet drug treatment (supplemental Figure 4).
Increased ADP- and epi-induced platelet aggregation was strongly related to atrial fibrillation (PR > 1, all group comparisons; Figure 2A-B; supplemental Figure 5), whereas dyslipidemia was associated with ADP high-responsive platelets (Figure 2A; supplemental Figure 5A) and venous thromboembolism (VTE) with epi high-responsive platelets in the LTA (P < .05; Figure 2B; supplemental Figure 5B). Based on the analysis of 56 common variants related to platelet phenotype and function (supplemental Table 2), the variants rs1671152 and rs1613662 in glycoprotein 6 (GP6), encoding the platelet lineage–specific collagen receptor GPVI, demonstrated predictive impact for increased platelet ADP aggregability (Figure 2C; supplemental Figure 6A), whereas the gain-of-function variant rs3737224 in platelet endothelial aggregation receptor-1 (PEAR1) and the megakaryocyte and platelet inhibitory receptor G6b (MPIG6B) variant rs11575845 showed predictive impact for platelet high reactivity to epi (all P < .05; Figure 2D; supplemental Figure 6B).
Predictive value of clinical factors and platelet-related common genetic variants for platelet high responsiveness to ADP and epinephrine. Associations between platelet high aggregability induced by a low-response concentration (0.5 μM) of ADP and/or epi and clinical characteristics (A-B), platelet–related common genetic variants (C-D). The group comparisons, (A,C) ADP epi and (B,D) epi, were investigated using Poisson regressions with robust standard errors. The model was adjusted for age, sex, platelet count (C-D), plus platelet function-interfering medications (A-B). Alt, alternative allele; MI, myocardial infarction; Ref, reference allele.
Predictive value of clinical factors and platelet-related common genetic variants for platelet high responsiveness to ADP and epinephrine. Associations between platelet high aggregability induced by a low-response concentration (0.5 μM) of ADP and/or epi and clinical characteristics (A-B), platelet–related common genetic variants (C-D). The group comparisons, (A,C) ADP epi and (B,D) epi, were investigated using Poisson regressions with robust standard errors. The model was adjusted for age, sex, platelet count (C-D), plus platelet function-interfering medications (A-B). Alt, alternative allele; MI, myocardial infarction; Ref, reference allele.
Our observation that platelet ADP and/or epi HR were associated with nonagonist-specific elevated platelet aggregation is consistent with previous studies,9,10 supporting ADP and epi as important confounding agonists, which might also contribute to high on-treatment platelet reactivity.7,8 Our flow cytometric data suggest that high-reactive platelets to ADP/epi are not associated with preactivated platelets in vivo and that tissue factor deruved from other cell types, likely via extracellular vesicles, interact with unstimulated or positively primed platelets through noncanonical adhesion pathways.11
The strong association between the prevalence of atrial fibrillation and enhanced ADP and epi aggregation coincides with previous small cohort studies.12,13 Also, the relation between dyslipidemia and high platelet aggregation to ADP supports reports studying platelets from hyperlipidemic animals and humans.14,15 Notably, we found that the history of VTE predicts high-responsive platelets preferentially to epi. However, no significant relationship was observed for other cardiovascular diseases, risk factors, and comorbidities as described for the sticky platelet syndrome.6
Eight genes were related to platelet ADP and/or epi aggregability, including cell surface receptors (GP6, PEAR1, MPIG6B, and P2RY12), serine/threonine protein kinases (AKT2 and MAP2K2), regulators of Gα/i protein–coupled receptors (RGS4; RGS5), and microRNA-host gene (MIR100G).16 The nonsynonymous variants rs1613662 (G/A) and rs1671152 (T/G) in GP6 are well known to be associated with higher sensitivity to GPVI–induced platelet activation17-19 but hitherto have been unreported to be related to platelet ADP hyperfunction in Western Europeans. The platelet endothelial aggregation receptor, PEAR1 contributes to platelet activation via Src family kinase–mediated signaling through αIIbβ3-(in)dependent platelet-platelet contacts.20,21 The synonymous PEAR1 variant rs3737224 demonstrated predictive potential for enhanced platelet aggregability in response to epi rather than to ADP, which was not reported before.19MPIG6B encodes the specific megakaryocyte and platelet inhibitory receptor G6b-B, a member of the immunoglobulin superfamily, which binds the tandem tyrosine phosphatases Shp2 and Shp1 after phosphorylation of tyrosine sites within its 2 immunoreceptor tyrosine-based inhibitory motifs.22 Antibody-mediated crosslinking of this inhibitory receptor suppressed Ca2+–independent platelet activation in response to ADP and a GPVI-agonist.23 However, the predictive impact of the MPIG6B variant rs11575845 on increased epi–induced platelet aggregation was not demonstrated yet.
Nevertheless, this association study, based on the analysis of single-day blood samples, needs confirmation for causal relations and mechanistic explanations. Further limitations include missing flow cytometric agonist–induced platelet function data, no adjustment for multiple testing and for other analyzed genes, and a lack of data validation with a second cohort.
Overall, this study identifies common and specific cardiovascular and genetic determinants of platelet ADP and epi HR independent of established confounders. Furthermore, our data provide new potential insights into the interrelation of ADP/epi–dependent platelet functions with noncanonical platelet receptors, that is, GPVI and G6b-B. Evaluation of platelet ADP and epi HR might provide (1) predictive value for enhanced platelet activity in atrial fibrillation, dyslipidemia, and VTE and (2) benefits for personalized antiplatelet treatment.
The protocol and sampling strategy for the study were approved by the local ethics committee (837.020.07), and the study was planned according to the principles of the revised Helsinki protocol. The local and state data safety commissioners also approved the sampling plan. Each participant provided informed consent before the study.
Acknowledgments: The Gutenberg Health Study is funded through the Government of Rhineland-Palatinate (Stiftung Rheinland-Pfalz fur Innovation, contract AZ 961-386261/733), the research programs Wissenschaft Zukunft and Center for Translational Vascular Biology of the Johannes Gutenberg University of Mainz, and its contract with Boehringer Ingelheim and PHILIPS Medical Systems, including an unrestricted grant for the Gutenberg Health Study. G.B. was supported by a PhD fellowship from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant (agreement no. 813409).
Contribution: K.J., M.P.-N., and P.S.W. conceived the study; B.D., G.B., and S.v.U.-S. performed platelet function analysis; K.J., V.t.C., A.G., and P.S.W. designed the statistical analysis plan; A.G. conducted the statistical analysis; G.B., S.v.U.S., and K.J. drafted the manuscript; and all authors participated in the interpretation of the findings, reviewed the manuscript, revised it critically before submission, and approved the final version of the manuscript.
Conflict-of-interest disclosure: H.t.C. received research funding outside the present study from Bayer; and received honoraria for consultation and/or advisory board participation from Bayer, Alveron, Galapagos, Portola, and Alexion, outside the present study. All reimbursements were transferred to the Cardiovascular Research Institute Maastricht. H.t.C. is a shareholder with Coagulation Profile, a university spinoff, small diagnostic company not involved in the present study. P.S.W. has received research funding outside the present study from Boehringer Ingelheim, Sanofi-Aventis, Bayer Healthcare, Daiichi Sankyo Europe, and Novartis; and received outside the present study honoraria for lectures or consulting from Boehringer Ingelheim, Bayer HealthCare, Evonik, AstraZeneca, and Sanofi-Aventis. P.S.W. is a principal investigator of the data-independent acquisition-based systems medicine (DIASyM) research core (BMBF 161L0217A). The remaining authors declare no competing financial interests.
Correspondence: Kerstin Jurk, Center for Thrombosis and Hemostasis, University Medical Center Mainz of the Johannes Gutenberg-University Mainz, Langenbeckstr. 1, Mainz 55131, Germany; email: kerstin.jurk@unimedizin-mainz.de.
References
Author notes
The R-code used for the analyses is available upon request. Based on the written consent of the study participants, the data will only be made available locally within the framework of the access procedure specified in the rules of procedure of the research consortium. Interested researchers should direct their inquiries to the Gutenberg Health Study Steering Committee via the coordinating principal investigator (contact via info@ghs-mainz.de).
The full-text version of this article contains a data supplement.