TO THE EDITOR:
NUP98, a nuclear pore complex component, functions as abnormal transcription factor in fusion proteins, and its partner genes in NUP98 fusions also possess transcription-related domains.1 Acute myeloid leukemia (AML) with NUP98 rearrangement (NUP98r) is a newly introduced diagnostic entity in both the fifth edition of the World Health Organization (WHO) classification2 and the 2022 International Consensus Classification (ICC) guidelines.3 In the WHO fifth edition classification, it is presented as a distinct diagnostic entity under the classification AML with NUP98r.2 In the ICC 2022 guidelines, it is described under the category AML with other rare recurring translocations, specifically as AML with NUP98::NSD1, NUP98::KMD5A, and NUP98 and other partners.3 Notably, the ICC 2022 guidelines elaborate that these 3 diagnoses are predominantly observed in infants and children. In pediatric cases, NUP98 translocations have been reported in ∼3.8% of patients and are associated with poor prognosis.4 Conversely, there is limited study concerning the prevalence and treatment outcomes related to NUP98r in adults.
In this study, we aimed to comprehensively analyze the relatively understudied prevalence of NUP98r in adults and discuss potential therapeutic strategies. The institutional review board of Yonsei University Health System, Severance Hospital, Seoul, Korea, approved this study and waived the need for informed consent.
In a single institution, from August 2018 to March 2023, newly diagnosed adult patients (aged ≥18 years) with AML who were identified with NUP98r through targeted RNA sequencing were investigated, as well as those who underwent anticancer treatment.
The G-banding karyotyping procedure followed standard protocols on heparinized bone marrow aspirate. Targeted RNA sequencing was performed on total RNA extracted from bone marrow aspirate using QIAamp RNA Blood Mini Kit (Qiagen, Hilden, Germany). A complementary DNA library was prepared using the Archer FusionPlex Pan-Heme kit (ArcherDX, Boulder, CO), which contains gene-specific primers targeting 199 genes and universal primers designed to detect unknown fusion partners even if only 1 fusion partner is included in the target (supplemental Table 1).5 For mutation detection, targeted DNA sequencing was performed using a customized set of probes (Dxome Co Ltd, Seongnam-si, Korea) targeting 497 genes (supplemental Table 2).6 Targeted RNA and DNA sequencings were done with the NextSeq 550Dx instrument (Illumina, San Diego, CA). FLT3-internal tandem duplication (ITD) was confirmed and quantified using fragment analysis according to the previously described method.7
For our study, 351 adult patients were newly diagnosed with AML, with 260 undergoing targeted RNA sequencing. Among the 260 patients, NUP98r was diagnosed in 13 (5.0%), with an average fusion read count of 1141× (range, 260-2873) and fusion read percentage of 24.2% (range, 4.0-54.8), and 4 cases were confirmed by reverse transcript polymerase chain reaction. Of these patients, 10 received anticancer treatment at our institution, whereas 3 patients were transferred or declined treatment. During the same period, 34 pediatric patients with AML were diagnosed, and targeted RNA sequencing was performed on 29 of them; however, none were diagnosed with NUP98r.
The demographics for the 10 patients are described in Table 1. Their average age was 42.7 years, indicating a relatively younger age group within the cohort of patient with AML.8 Among these patients, 2 older individuals were treated with hypomethylating agent and/or venetoclax, but they passed away during cancer treatment. The remaining 8 patients underwent conventional induction chemotherapy (7-day cytarabine plus 3-day anthracycline). Among them, 4 patients with an FLT3 mutation received FLT3 inhibitors, such as midostaurin. After standard 7 + 3 induction chemotherapy, 6 patients achieved complete remission (CR), a rate of 75.0%, consistent with rates reported in the literature for AML. Two patients who did not achieve CR through the standard 7 + 3 regimen achieved CR after receiving cladribine-based reinduction chemotherapy. In addition, all patients underwent allogeneic hematopoietic stem cell transplantation (HSCT). One patient experienced relapse and subsequently underwent a second allogeneic HSCT, whereas the remaining patients did not experience relapse. Among the 8 patients, including the 1 who received a second allogeneic HSCT, 6 are currently in CR, 1 succumbed to COVID-19, and 1 was lost to follow-up after 2 years and 4 months. The median follow-up duration after diagnosis in patients who received transplantation was 39 months (range, 11-53).
Patient demographics
| Patient no. . | Sex . | Age at diagnosis, y . | Gene rearrangement . | Chromosome karyotyping . | Mutation analysis (VAF or allele ratio∗) . | Chemotherapy or conditioning regimen† . | Donor type (HLA matching, HLA-A, -B, -C, -DRB1, and -DQB1) . | Disease status at HSCT . | Relapse after HSCT/d . | F/U . | Outcome . | Cause of death . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | M | 65 | NUP98::HMGB3 | 47,XY,inv(9)(p11q13),+21[20] | FLT3-ITD (0.18) | Dec | 1 y 2 mo | Death | Leukemia | |||
| P2 | M | 74 | NUP98::NSD1 | 47,XY,+8[17]/46,XY,del(5)(q22q35)[3] | FLT3-ITD (1.95) FLT3 (9.2%) DNMT3A (42.4%) | Dec, Aza + Ven | 2 mo | Death | Leukemia | |||
| P3 | F | 57 | NUP98::DDX10 | 46,XX,inv(11)(p15q22)[9]/46,XX[12] | None | Bu + Flu + ATG | MUD (10/10) | CR | No | 11 mo | Death | COVID-19 |
| P4 | F | 33 | NUP98::TOP1 | 46,XX,del(9)(q13q31),t(11;20)(p15;q11)[17]/46,idem,del(7)(q32)[3]/46,XX[2] | RAD21 (40.3%) | Bu + Cy + ATG | MUD (9/10) | CR | No | 3 y 10 mo | Alive | |
| P5 | M | 23 | NUP98::NSD1 | 46,XY[20] | FLT3-ITD (0.72) RUNX1 (46.7%) | Flu + Cy + TBI | Haplo | CR | No | 4 y 5 mo | Alive | |
| P6 | M | 34 | NUP98::NSD1 | 46,XY[20] | FLT3-ITD (0.75) | Bu + Cy + ATG | MUD (10/10) | CR | No | 4 y 3 mo | Alive | |
| P7 | F | 20 | NUP98::HOXA9 | 46,XX,t(7;11)(p15;p15)[20] | NRAS (15.3%) | Flu + Cy + TBI | Haplo | CR | Yes/1 y | 4 y 5 mo | Alive after 2nd HSCT | |
| P8 | F | 58 | NUP98::HOXC13 | 46,XX,t(11;12)(p15;q13)[20] | FLT3-ITD (0.26) PTPN11 (6.4%) NRAS (6.3%) | Bu + Flu + ATG | MUD (10/10) | CR | No | 2 y 4 mo | F/U lost | |
| P9 | M | 37 | NUP98::PHF23 | 46,XY,der(16)t(3;16)(q21;p13.1),del(17)(p11.2)[20] | TP53 (WGD) CREBBP (WGD) AXIN1 (WGD) NRAS (27.3%) NRAS (12.3%) | Bu + Cy + ATG | MUD (10/10) | CR | No | 1 y 5 mo | Alive | |
| P10 | F | 26 | NUP98::NSD1 | 46,XX,t(5;12)(q35;q24.1)?c[20] | FLT3-ITD (0.39) WT1 (46.5%) | Bu + Cy + ATG | MSD | CR | No | 2 y 7 mo | Alive |
| Patient no. . | Sex . | Age at diagnosis, y . | Gene rearrangement . | Chromosome karyotyping . | Mutation analysis (VAF or allele ratio∗) . | Chemotherapy or conditioning regimen† . | Donor type (HLA matching, HLA-A, -B, -C, -DRB1, and -DQB1) . | Disease status at HSCT . | Relapse after HSCT/d . | F/U . | Outcome . | Cause of death . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | M | 65 | NUP98::HMGB3 | 47,XY,inv(9)(p11q13),+21[20] | FLT3-ITD (0.18) | Dec | 1 y 2 mo | Death | Leukemia | |||
| P2 | M | 74 | NUP98::NSD1 | 47,XY,+8[17]/46,XY,del(5)(q22q35)[3] | FLT3-ITD (1.95) FLT3 (9.2%) DNMT3A (42.4%) | Dec, Aza + Ven | 2 mo | Death | Leukemia | |||
| P3 | F | 57 | NUP98::DDX10 | 46,XX,inv(11)(p15q22)[9]/46,XX[12] | None | Bu + Flu + ATG | MUD (10/10) | CR | No | 11 mo | Death | COVID-19 |
| P4 | F | 33 | NUP98::TOP1 | 46,XX,del(9)(q13q31),t(11;20)(p15;q11)[17]/46,idem,del(7)(q32)[3]/46,XX[2] | RAD21 (40.3%) | Bu + Cy + ATG | MUD (9/10) | CR | No | 3 y 10 mo | Alive | |
| P5 | M | 23 | NUP98::NSD1 | 46,XY[20] | FLT3-ITD (0.72) RUNX1 (46.7%) | Flu + Cy + TBI | Haplo | CR | No | 4 y 5 mo | Alive | |
| P6 | M | 34 | NUP98::NSD1 | 46,XY[20] | FLT3-ITD (0.75) | Bu + Cy + ATG | MUD (10/10) | CR | No | 4 y 3 mo | Alive | |
| P7 | F | 20 | NUP98::HOXA9 | 46,XX,t(7;11)(p15;p15)[20] | NRAS (15.3%) | Flu + Cy + TBI | Haplo | CR | Yes/1 y | 4 y 5 mo | Alive after 2nd HSCT | |
| P8 | F | 58 | NUP98::HOXC13 | 46,XX,t(11;12)(p15;q13)[20] | FLT3-ITD (0.26) PTPN11 (6.4%) NRAS (6.3%) | Bu + Flu + ATG | MUD (10/10) | CR | No | 2 y 4 mo | F/U lost | |
| P9 | M | 37 | NUP98::PHF23 | 46,XY,der(16)t(3;16)(q21;p13.1),del(17)(p11.2)[20] | TP53 (WGD) CREBBP (WGD) AXIN1 (WGD) NRAS (27.3%) NRAS (12.3%) | Bu + Cy + ATG | MUD (10/10) | CR | No | 1 y 5 mo | Alive | |
| P10 | F | 26 | NUP98::NSD1 | 46,XX,t(5;12)(q35;q24.1)?c[20] | FLT3-ITD (0.39) WT1 (46.5%) | Bu + Cy + ATG | MSD | CR | No | 2 y 7 mo | Alive |
ATG, antithymocyte globulin; Aza, azacitidine; Bu, busulfan; Cy, cyclophosphamide; Dec, decitabine; F, female; F/U, follow-up; Flu, fludarabine; Haplo, haploidentical donor; M, male; MSD, matched sibling donor; MUD, matched unrelated donor; TBI, total body irradiation; VAF, variant allele frequency; Ven, venetoclax; WGD, whole-gene deletion.
VAF (%) was calculated from next-generation sequencing as a percentage by dividing the number of variant allele reads at each position by the total number of reads at that position. FLT3-ITD allele ratio was calculated from fragment analysis as the ratio of the mutant product's peak height to the wild-type product's peak height.
We presented chemotherapy regimens for individuals who had not undergone transplantation (P1 and P2) and conditioning regimens for the remaining patients who had undergone HSCT.
We identified high prevalence of NUP98r in adult patients with AML (5.0%) using targeted RNA sequencing. NUP98::NSD1 occurred in 4 cases (2.3%), whereas the remaining 6 cases exhibited various other rearrangements. Consequently, we believe that NUP98r in adults has been underrepresented and is underexplored. Similar to the case with core-binding factor AML,9 there may be a higher prevalence of NUP98r in the Asian population. For instance, it has been speculated that NUP98::HOXA9 occurs more frequently in Asians,1 and there have been reports of NUP98r with a 2.7% prevalence in Asians.10
NUP98, located at the terminal (p15.5) of chromosome 11, is associated with cryptic translocations.11 We were able to identify only 4 cases of translocations or inversions involving chromosome 11. Therefore, for a precise diagnosis of AML, it is imperative to conduct NUP98-targeted analysis. The break points of NUP98 in our patients were located between exons 11 and 16, whereas those in a previous report were between exons 8 and 14.1 Therefore, it is anticipated that covering exons 8 through 16 of a total of 33 exons of NUP98 should be sufficient. For targeted DNA sequencing, FLT3-ITD was found in 6 patients (60%) and NRAS in 3 patients (30%), consistent with prior reports emphasizing high frequencies of FLT3-ITD.4,12 However, the prognostic significance of FLT3-ITD in comparison with patients without this disorder has not been conclusively established in this study, suggesting a need for further investigation of the existing literature.12,13
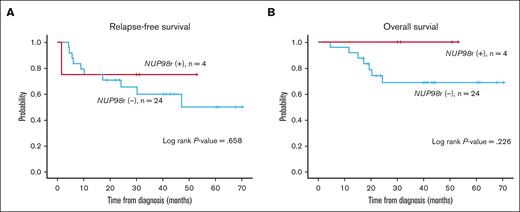
In contrast to previous studies that have indicated a poor prognosis for AML with NUP98r,4 we offer a different perspective. Eight patients who underwent induction chemotherapy ultimately proceeded to allogeneic HSCT, and 7 did not experience relapse. At our institution, we adopted a rapid HSCT approach, with allogeneic HSCT performed as early as 3 months after diagnosis. This intervention aligns with previous research, suggesting that prompt transplantation enhances survival rates in this condition.14 The previous study by Shen Y et al involved only 13 patients who were eligible for transplanatation.14 Still, we presented all patients with NUP98r regardless of HSCT eligibility, and patients who were unable to undergo transplantation showed a dismal outcome. Furthermore, there was no difference in relapse-free survival and overall survival between NUP98r+ and NUP98r− cases among FLT3-ITD+ patients with AML who underwent HSCT at our institution (Figure 1). Meanwhile, ongoing research on the effectiveness of menin inhibitors as potential therapeutics for NUP98-rearranged leukemia shows promising results.15,16 These novel treatments may further improve the prognosis of NUP98-rearranged patients.
Survival outcomes of FLT3-ITD+ AML according to NUP98r status among those who received allogeneic HSCT (n = 28). (A-B) The Kaplan-Meier plots of relapse-free survival (RFS) (A) and overall survival (OS) (B) in NUP98r+ and NUP98r− patients. Statistical analysis was done with IBM SPSS Statistics software version 28.0.0.0. P values <.05 were considered statistically significant. RFS was determined from the time of AML diagnosis to the first relapse, death, or last follow-up. OS was measured from the time of initial diagnosis to death from any cause or last follow-up.
Survival outcomes of FLT3-ITD+ AML according to NUP98r status among those who received allogeneic HSCT (n = 28). (A-B) The Kaplan-Meier plots of relapse-free survival (RFS) (A) and overall survival (OS) (B) in NUP98r+ and NUP98r− patients. Statistical analysis was done with IBM SPSS Statistics software version 28.0.0.0. P values <.05 were considered statistically significant. RFS was determined from the time of AML diagnosis to the first relapse, death, or last follow-up. OS was measured from the time of initial diagnosis to death from any cause or last follow-up.
This study had several limitations. Because this was a single-center study, generalizability of the results may be limited. Furthermore, its small sample size may affect our ability to reach definitive conclusions. Our findings must be verified through large-scale multicenter studies in the future.
In conclusion, we found that NUP98r occur in adults at a rate of 5.0%, which is comparable or higher than the rate in pediatric cases. Additionally, we observed encouraging outcomes with a treatment strategy that includes induction chemotherapy followed by allogeneic HSCT, in accordance with the concept of rapid transplantation to improve survival rates.
Acknowledgment: This work was supported by the National Research Foundation of Korea (NRF-2021R1I1A1A01045980).
Contribution: N.K. and S.S. conceptualized the study; S.-T.L. and J.R.C. designed the methodology; N.K. was responsible for investigation; H. Cho., J.E.J., J.-W.C., and H. Chung provided the resources; N.K. and S.S. curated the data; N.K. wrote the original draft; Y.J.C., S.S., and H. Chung reviewed and edited the manuscript; S.-T.L., J.S., and J.R.C. supervised the study; and all authors have read and agreed to the published version of the manuscript.
Conflict-of-interest disclosure: S.-T.L. and J.R.C. are employed by Dxome. The remaining authors declare no competing financial interests.
Correspondence: Haerim Chung, Division of Hematology, Department of Internal Medicine, Yonsei University College of Medicine, Severance Hospital, 50 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; email: SANHO23@yuhs.ac; and Saeam Shin, Department of Laboratory Medicine, Yonsei University College of Medicine, Severance Hospital, 50 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea; email: saeam0304@yuhs.ac.
References
Author notes
The data sets generated during and/or analyzed during the current study are available on reasonable request from the corresponding authors, Haerim Chung (SANHO23@yuhs.ac) and Saeam Shin (saeam0304@yuhs.ac).
The full-text version of this article contains a data supplement.