Key Points
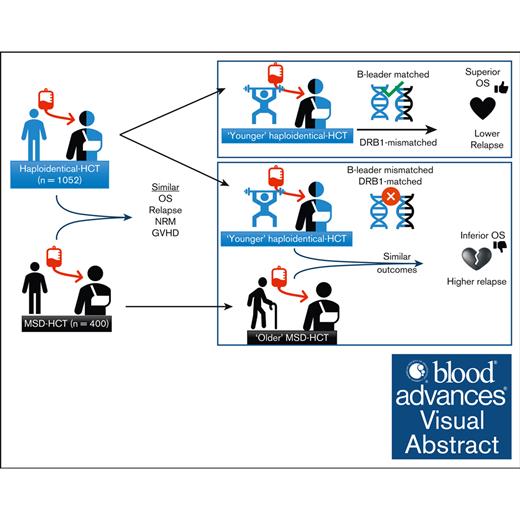
Haploidentical-HCT, especially with younger donors, may offer comparable outcomes to MSD-PTCy HCT.
In older patients, younger haploidentical B-leader–matched donors HCT had superior OS than older MSDs.
Visual Abstract
HLA-matched sibling donors (MSDs) are preferred for hematopoietic cell transplantation (HCT). However, the use of alternative donors, especially haploidentical, is increasing, as is our understanding of the impact of HLA factors such as B-leader and DRB1-matching on its outcomes. Yet, data comparing these donor types, particularly considering these HLA factors, is lacking. Herein, we compared haploidentical-HCT (n = 1052) with MSD-HCT (n = 400), both with posttransplant cyclophosphamide (PTCy)-based graft-versus-host disease prophylaxis. In multivariate analysis, haploidentical group had similar overall survival (OS; hazard ratio (HR), 0.94; 95% confidence interval [CI], 0.78-1.14; P = .54), nonrelapse mortality (HR, 0.98; 95% CI, 0.72-1.32; P = .87), and relapse (HR, 0.87; 95% CI, 0.70-1.08; P = .20) as the MSD group. Younger donor age was a significant predictor of improved OS. Next, we directly compared the outcomes of “younger” haploidentical (donor age <35 years, n = 347) vs an “older” MSD (donor age ≥50 years, n = 143) in older recipients (patient age ≥50 years). Patients with younger haploidentical B-leader–matched donors had significantly superior OS (HR, 0.65; 95% CI, 0.48-0.90; P = .009) than the older MSD group. Additionally, patients with younger DRB1-mismatched haploidentical donors (HR, 0.63; 95% CI, 0.46-0.87; P = .004) had significantly lower risk of relapse than older MSDs. Our study suggests that haploidentical-HCT may offer comparable outcomes to MSD-PTCy HCT. Moreover, among older patients, a younger haploidentical B-leader–matched donor might be preferable to an older MSD. These findings need validation in larger data sets.
Introduction
HLA matching is 1 of the most important determinants of hematopoietic cell transplantation (HCT) outcomes.1 Generally, an HLA-matched sibling donor (MSD) is considered the gold standard. However, haploidentical donor HCT is becoming increasingly common in the United States, surpassing MSD-HCT since 2020.2 Despite this rising trend, there is a dearth of data directly comparing these 2 donor types. Although the report from the Center for International Blood and Marrow Transplant Research suggests narrow differences in overall survival (OS) after MSD-HCT and haploidentical HCT, a definitive conclusion cannot be drawn from these unadjusted data.2 For instance, among patients with acute myeloid leukemia (AML) in first complete remission (CR1), the 3-year OS probability was 57% after MSD-HCT and 50% with haploidentical-HCT. Similar trends were observed in CR2+ (53% for both) and relapsed/persistent disease (31% vs 27%, respectively).2
One might question why a haploidentical donor would be considered when a MSD is available. A formal comparison between the 2 donor types is particularly relevant considering the advancements in optimal haploidentical donor selection, which have led to a deeper understanding of how certain HLA factors determine transplant outcomes. A recent study demonstrated that haploidentical donor-recipient pairs with HLA B-leader matching had significantly improved OS and lower nonrelapse mortality (NRM) compared with those who were B-leader mismatched.3 Also, HLA-DRB1 mismatching was associated with a lower risk of relapse and improved progression-free survival than DRB1-matched cases.3 This highlights the need for direct comparative studies to assess outcomes after MSD-HCT and haploidentical-HCT, considering both HLA and non-HLA factors.
Among non-HLA factors, donor age is a well-established predictor of survival.4-6 Evidence suggests “younger” (aged <35 years) donors might mitigate the negative effects of HLA-mismatching.7 This poses a common clinical dilemma: whether to use an “older” (aged ≥50 years) MSD or a younger alternative donor, especially in older patients for whom a sibling is also likely to be older. Although some studies showed similar or even better outcomes with a younger HLA-matched unrelated donor compared with older MSD,8-10 the potential benefits of a younger haploidentical donor compared with an older MSD remain unclear.
Therefore, we pursued this study with 2 main objectives. First, we compared the outcomes of haploidentical-HCT vs MSD-HCT with a hypothesis that haploidentical-HCT would achieve similar OS rates to MSD-HCT. Second, we investigated whether a younger, particularly B-leader matched, haploidentical donor could offer superior OS than an older MSD.
Methods
We used 2 publicly available Center for International Blood and Marrow Transplant Research data sets11 from the referenced publications.3,12 The local institutional review board (FHIRB0020181) approved the study, which was conducted in accordance with the Declaration of Helsinki. Our study population included patients with AML, acute lymphoblastic leukemia, or myelodysplastic neoplasm (MDS) who underwent a haploidentical or MSD-HCT between 2008 and 2017. All patients received posttransplant cyclophosphamide (PTCy)–based graft-versus-host disease (GVHD) prophylaxis.
Statistical methods
Our first aim was to compare the entire haploidentical cohort with the entire MSD cohort. However, such a retrospective analysis, especially when comparing a partially matched donor approach (haploidentical) with MSD (considered the gold standard), may be susceptible to underlying selection biases, center-specific preferences, and baseline patient characteristic differences. Standard multivariable regression models might not fully account for these factors. Therefore, we adopted a quasi-experimental technique of “doubly robust” analysis,13 which combines the propensity score–based methods with multivariable regression models adjusting for baseline characteristics and center-effect for a more robust analysis. We first used a machine learning method of “gradient boosting model,” which is implemented in the R package “twang”14 to estimate the propensity scores (likelihood of receiving a haploidentical donor). Then, we performed propensity score matching on patient age (<50 vs ≥50 years), disease (acute leukemia vs MDS), disease stage (early-intermediate vs advanced), graft type (bone marrow vs peripheral blood), conditioning intensity (myeloablative vs nonmyeloablative/reduced intensity), HCT comorbidity index score (0-2 vs ≥3) and Karnofsky performance scale score (<90 vs ≥90) using the “MatchIt” R package.15
The resulting matched-pair cohort included 1052 haploidentical patients and 400 patients in the MSD group. Given that B-leader matching (affecting OS and NRM) and DRB1-matching (influencing relapse) in the haploidentical group are known to affect outcomes, we conducted preplanned subgroup analyses in which the haploidentical group was categorized on these HLA factors. Because neither B-leader nor DRB1 matching was a predictor of GVHD,3 the entire haploidentical group was compared with the MSD group for GVHD outcomes. To strengthen our methods, we additionally performed inverse probability of treatment weighting (IPTW) analyses on the entire cohort for OS, NRM, relapse. IPTW assigns weights to patients based on their likelihood of receiving a haploidentical transplant, accounting for any imbalances. These analyses complement propensity score matching, potentially leading to more robust estimates by using all available data.
Our second aim was to compare a younger haploidentical donor to an older MSD. Due to a strong correlation between donor age and recipient age, particularly in the MSD group, this analysis was restricted to older patients (≥50 years). Within this cohort, we categorized donors as younger vs older based on a series of 2-sample t tests and the receiver operating characteristic curve analysis. These analyses identified distinct age thresholds for MSD (∼45-50 years) and haploidentical donors (∼30-35 years). Based on these cutoffs, 2 groups were created: younger haploidentical (donor age < 35 years, n = 347) and older MSD (donor age ≥ 50 years, n = 143). Supplemental Figure 1 details the initial sample sizes and sample sizes of the respective groups.
Statistical analysis methods for both aims
Baseline characteristics between the groups were summarized using descriptive statistics with median and interquartile range for continuous variables and numbers with percentages for categorical variables. These were compared between the groups using the χ2 test for categorical variables, and the Mann-Whitney U test for continuous variables. Our primary outcome of interest was OS. Secondary outcomes included NRM, relapse, overall chronic GVHD and grade 2 to 4 acute GVHD. In univariate analysis, the Kaplan-Meier method was used to estimate OS probabilities. The cumulative incidence method accounting for competing risks was used to estimate the incidences of relapse, NRM, and GVHD. Competing risks considered were death for GVHD; disease relapse for NRM; and death before relapse (NRM) for relapse. The median follow-up of the MSD group was shorter (∼24 months) than the haploidentical group (∼32 months). Therefore, the probabilities of outcomes at 24 months are reported for all outcomes, except for acute GVHD, which is at day 100. Multivariable analysis was performed using Cox proportional hazards (PH) models. Cause-specific hazards are reported for all outcomes with competing events. All analyses were adjusted for center effect using the shared-frailty model. Because of testing of multiple outcomes, a 2-sided P value of < .01 was considered significant. We tested the PH assumption for all variables graphically and by including interaction terms with time in the model. Interaction effects between the main effect (donor type) and other statistically significant covariables were tested and none were noted in multivariable regression analyses. We noted that race violated the PH assumptions for the outcome of acute GVHD and stage violated the PH assumptions for the outcome of relapse. Consequently, we stratified the multivariate analysis for acute GVHD by race and for relapse by disease stage. All analyses were done using SAS version 9.4 TS Level 1M8, and R Statistical Software (version 4.3.1; R Core Team 2023).
Results
Haploidentical vs MSD: propensity score matched–cohort analysis
The haploidentical group included slightly older patients (median age, 49.1 vs 46.2 years; P = .03) with younger donors (median age, 39.1 vs 44.7 years; P < .0001) compared with the MSD group. Other baseline characteristics were similar (Table 1). Most patients had acute leukemia (82%-83%), early/intermediate disease (71%-72%), received myeloablative conditioning (53%-55%), and peripheral blood graft (64%-68%). Karnofsky performance scale score and HCT comorbidity index score were also similar, with most patients having a score of ≥90 (57%-58%) and 0 to 2 (53%-57%), respectively. The haploidentical-PTCy group had a significantly longer median follow-up (32.3 months) than the MSD-PTCy group (24.4 months).
Baseline characteristics
| . | Haplo-PTCy . | MSD-PTCy . | P . | Younger haplo-PTCy . | Older MSD-PTCy . | P . |
|---|---|---|---|---|---|---|
| n = 1052 . | n = 400 . | n = 347 . | n = 143 . | |||
| Patient age, y | .12 | - | ||||
| <50 | 539 (51.2) | 223 (55.8) | 347 (100) | 143 (100) | ||
| ≥50 | 513 (48.8) | 177 (44.2) | ||||
| Median (IQR) | 49.1 (31.7-63.3) | 46.2 (31.2-59.5) | .03 | 59.4 (55.1-64) | 61.2 (56.6-66) | .02 |
| Donor age | .11 | |||||
| Median (IQR) | 39.1 (29.7-47.6) | 44.7 (30-56.3) | <.0001 | 28.1 (24.1-31.5) | 58.9 (54.9-63.5) | <.0001 |
| Sex | .66 | .71 | ||||
| Male | 623 (59.2) | 242 (60.5) | 217 (62.5) | 92 (64.3) | ||
| Female | 429 (40.8) | 158 (39.5) | 130 (37.5) | 51 (35.7) | ||
| Disease | .47 | .06 | ||||
| AML/ALL | 877 (83.4) | 327 (81.8) | 260 (74.9) | 95 (66.4) | ||
| MDS | 175 (16.6) | 73 (18.2) | 87 (25.1) | 48 (33.6) | ||
| Disease stage | .79 | .35 | ||||
| Early-intermediate | 749 (71.2) | 288 (72) | 240 (69.2) | 92 (64.3) | ||
| Advanced | 296 (28.1) | 110 (27.5) | 107 (30.8) | 50 (35) | ||
| Missing | 7 (0.7) | 2 (0.5) | - | 1 (0.7) | ||
| Conditioning | .48 | .19 | ||||
| MAC | 557 (53) | 220 (55) | 110 (31.7) | 54 (37.8) | ||
| RIC/NMA | 495 (47) | 180 (45) | 237 (68.3) | 89 (62.2) | ||
| Graft type | .17 | .28 | ||||
| PB | 672 (63.9) | 271 (67.8) | 235 (67.7) | 104 (72.7) | ||
| BM | 380 (36.1) | 129 (32.2) | 112 (32.3) | 39 (27.3) | ||
| KPS | .94 | .51 | ||||
| <90 | 431 (41) | 166 (41.5) | 168 (48.4) | 75 (52.4) | ||
| ≥90 | 594 (56.5) | 231 (57.8) | 171 (49.3) | 67 (46.8) | ||
| Missing | 27 (2.6) | 3 (0.8) | 8 (2.3) | 1 (0.7) | ||
| HCT-CI | .24 | .32 | ||||
| 0-2 | 558 (53) | 226 (56.5) | 170 (49) | 63 (44.1) | ||
| ≥3 | 494 (47) | 174 (43.5) | 177 (51) | 80 (55.9) | ||
| D/R CMV | .89 | .16 | ||||
| Pos/Pos | 475 (45.2) | 171 (42.8) | 138 (39.8) | 59 (41.3) | ||
| Pos/Neg | 92 (8.8) | 36 (9) | 22 (6.3) | 15 (10.5) | ||
| Neg/Pos | 280 (26.6) | 99 (24.8) | 111 (32) | 33 (23.1) | ||
| Neg/Neg | 198 (18.8) | 79 (19.8) | 75 (21.6) | 32 (22.4) | ||
| Missing | 7 (0.7) | 15 (3.8) | 1 (0.3) | 4 (2.8) | ||
| D/R sex | .75 | .24 | ||||
| Others | 815 (77.5) | 313 (78.2) | 274 (79) | 106 (74.1) | ||
| Fem-to-Male | 237 (22.5) | 87 (21.8) | 73 (21) | 37 (25.9) | ||
| Race | .79 | .94 | ||||
| White | 743 (70.6) | 279 (69.8) | 260 (74.9) | 109 (76.2) | ||
| Others | 244 (23.2) | 95 (23.8) | 75 (21.6) | 32 (22.4) | ||
| Missing | 65 (6.2) | 26 (6.4) | 12 (3.5) | 2 (1.4) | ||
| Time to HCT, median months (IQR) | 7 (4.5-14.5) | 6.7 (4.3-12.5) | .15 | 6.1 (4.2-10.9) | 6.7 (4.4-10.4) | .42 |
| Year of HCT, median (IQR) | 2016 (2015-2017) | 2016 (2015-2016) | <.0001 | 2016 (2015-2017) | 2015 (2015-2016) | <.0001 |
| Follow-up among survivors, median months (IQR) | 32.3 (24-43) | 24.4 (12.5-36.7) | <.0001 | 35.1 (24.1-39.7) | 24.1 (12.4-37) | .0019 |
| . | Haplo-PTCy . | MSD-PTCy . | P . | Younger haplo-PTCy . | Older MSD-PTCy . | P . |
|---|---|---|---|---|---|---|
| n = 1052 . | n = 400 . | n = 347 . | n = 143 . | |||
| Patient age, y | .12 | - | ||||
| <50 | 539 (51.2) | 223 (55.8) | 347 (100) | 143 (100) | ||
| ≥50 | 513 (48.8) | 177 (44.2) | ||||
| Median (IQR) | 49.1 (31.7-63.3) | 46.2 (31.2-59.5) | .03 | 59.4 (55.1-64) | 61.2 (56.6-66) | .02 |
| Donor age | .11 | |||||
| Median (IQR) | 39.1 (29.7-47.6) | 44.7 (30-56.3) | <.0001 | 28.1 (24.1-31.5) | 58.9 (54.9-63.5) | <.0001 |
| Sex | .66 | .71 | ||||
| Male | 623 (59.2) | 242 (60.5) | 217 (62.5) | 92 (64.3) | ||
| Female | 429 (40.8) | 158 (39.5) | 130 (37.5) | 51 (35.7) | ||
| Disease | .47 | .06 | ||||
| AML/ALL | 877 (83.4) | 327 (81.8) | 260 (74.9) | 95 (66.4) | ||
| MDS | 175 (16.6) | 73 (18.2) | 87 (25.1) | 48 (33.6) | ||
| Disease stage | .79 | .35 | ||||
| Early-intermediate | 749 (71.2) | 288 (72) | 240 (69.2) | 92 (64.3) | ||
| Advanced | 296 (28.1) | 110 (27.5) | 107 (30.8) | 50 (35) | ||
| Missing | 7 (0.7) | 2 (0.5) | - | 1 (0.7) | ||
| Conditioning | .48 | .19 | ||||
| MAC | 557 (53) | 220 (55) | 110 (31.7) | 54 (37.8) | ||
| RIC/NMA | 495 (47) | 180 (45) | 237 (68.3) | 89 (62.2) | ||
| Graft type | .17 | .28 | ||||
| PB | 672 (63.9) | 271 (67.8) | 235 (67.7) | 104 (72.7) | ||
| BM | 380 (36.1) | 129 (32.2) | 112 (32.3) | 39 (27.3) | ||
| KPS | .94 | .51 | ||||
| <90 | 431 (41) | 166 (41.5) | 168 (48.4) | 75 (52.4) | ||
| ≥90 | 594 (56.5) | 231 (57.8) | 171 (49.3) | 67 (46.8) | ||
| Missing | 27 (2.6) | 3 (0.8) | 8 (2.3) | 1 (0.7) | ||
| HCT-CI | .24 | .32 | ||||
| 0-2 | 558 (53) | 226 (56.5) | 170 (49) | 63 (44.1) | ||
| ≥3 | 494 (47) | 174 (43.5) | 177 (51) | 80 (55.9) | ||
| D/R CMV | .89 | .16 | ||||
| Pos/Pos | 475 (45.2) | 171 (42.8) | 138 (39.8) | 59 (41.3) | ||
| Pos/Neg | 92 (8.8) | 36 (9) | 22 (6.3) | 15 (10.5) | ||
| Neg/Pos | 280 (26.6) | 99 (24.8) | 111 (32) | 33 (23.1) | ||
| Neg/Neg | 198 (18.8) | 79 (19.8) | 75 (21.6) | 32 (22.4) | ||
| Missing | 7 (0.7) | 15 (3.8) | 1 (0.3) | 4 (2.8) | ||
| D/R sex | .75 | .24 | ||||
| Others | 815 (77.5) | 313 (78.2) | 274 (79) | 106 (74.1) | ||
| Fem-to-Male | 237 (22.5) | 87 (21.8) | 73 (21) | 37 (25.9) | ||
| Race | .79 | .94 | ||||
| White | 743 (70.6) | 279 (69.8) | 260 (74.9) | 109 (76.2) | ||
| Others | 244 (23.2) | 95 (23.8) | 75 (21.6) | 32 (22.4) | ||
| Missing | 65 (6.2) | 26 (6.4) | 12 (3.5) | 2 (1.4) | ||
| Time to HCT, median months (IQR) | 7 (4.5-14.5) | 6.7 (4.3-12.5) | .15 | 6.1 (4.2-10.9) | 6.7 (4.4-10.4) | .42 |
| Year of HCT, median (IQR) | 2016 (2015-2017) | 2016 (2015-2016) | <.0001 | 2016 (2015-2017) | 2015 (2015-2016) | <.0001 |
| Follow-up among survivors, median months (IQR) | 32.3 (24-43) | 24.4 (12.5-36.7) | <.0001 | 35.1 (24.1-39.7) | 24.1 (12.4-37) | .0019 |
ALL, acute lymphoblastic leukemia; BM, bone marrow; CMV, cytomegalovirus; D/R, donor/recipient; HCT-CI, HCT comorbidity index; Fem-to-Male, female donor to male recipient; Haplo, haploidentical; IQR, interquartile range; KPS, Karnofsky performance status score; MAC, myeloablative conditioning; Neg, negative; NMA; nonmyeloablative conditioning; PB, peripheral blood; Pos, positive; RIC, reduced-intensity conditioning.
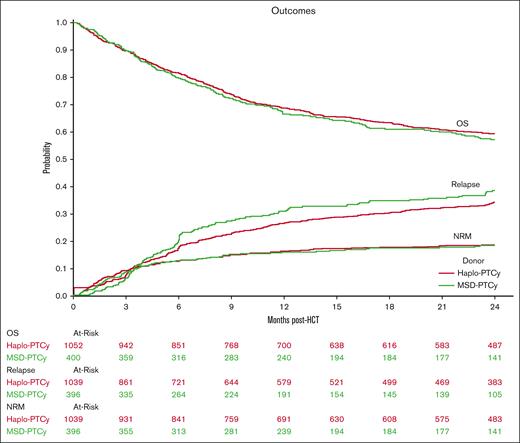
Results of the univariate analyses are presented in supplemental Table 1. Figure 1 displays Kaplan-Meier OS probability and cumulative incidence curves for relapse and NRM. Multivariate analysis of outcomes including OS, relapse, NRM, acute GVHD grades 2 to 4, and chronic GVHD showed similar results for the haploidentical-PTCy and MSD-PTCy groups (Table 2; supplemental Table 2). No significant differences in NRM, OS, or relapse were observed between the MSD group and the haploidentical groups categorized by B-leader matching or DRB1-matching. Similar findings were observed in the IPTW analyses (supplemental Table 3). There was no significant interaction between donor type and disease for relapse risk (haploidentical×acute lymphoblastic leukemia, P = .12; haploidentical×MDS, P = .56). However, because of potential heterogeneity in disease risks, we performed a sensitivity analysis restricted to patients with AML, which again showed similar results (supplemental Table 4).
Outcomes of haploidentical vs MSD-PTCy HCT. Kaplan-Meier probability of OS, and cumulative incidence of relapse and NRM comparing haploidentical donor (Haplo-PTCy, red) vs HLA-MSD (MSD-PTCy, green).
Outcomes of haploidentical vs MSD-PTCy HCT. Kaplan-Meier probability of OS, and cumulative incidence of relapse and NRM comparing haploidentical donor (Haplo-PTCy, red) vs HLA-MSD (MSD-PTCy, green).
Multivariate analysis: haploidentical vs MSD, propensity-matched cohort
| OS . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 400 | 177 | 57.2% (51.9-62.3) | Ref | ||
| Haplo-PTCy | 1052 | 471 | 59.3% (56.3-62.3) | 0.94 | 0.78-1.14 | .54 |
| MSD-PTCy | 400 | 177 | 57.2% (51.9-62.3) | Ref | ||
| Haplo: B-leader–matched | 635 | 275 | 61.7% (57.8-65.5) | 0.89 | 0.72-1.09 | .24 |
| Haplo: B-leader–mismatched | 417 | 196 | 55.8% (50.9-60.6) | 1.04 | 0.84-1.3 | .72 |
| OS . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 400 | 177 | 57.2% (51.9-62.3) | Ref | ||
| Haplo-PTCy | 1052 | 471 | 59.3% (56.3-62.3) | 0.94 | 0.78-1.14 | .54 |
| MSD-PTCy | 400 | 177 | 57.2% (51.9-62.3) | Ref | ||
| Haplo: B-leader–matched | 635 | 275 | 61.7% (57.8-65.5) | 0.89 | 0.72-1.09 | .24 |
| Haplo: B-leader–mismatched | 417 | 196 | 55.8% (50.9-60.6) | 1.04 | 0.84-1.3 | .72 |
| NRM . | N . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 396 | 73 | 19.9% (14.7-22.5) | Ref | ||
| Haplo-PTCy | 1039 | 203 | 12.1% (16.3-21.0) | 0.98 | 0.72-1.32 | .87 |
| MSD-PTCy | 396 | 73 | 18.5% (14.7-22.5) | Ref | ||
| Haplo: B-leader–matched | 627 | 109 | 16.5% (13.7-19.5) | 0.83 | 0.6-1.15 | .27 |
| Haplo: B-leader–mismatched | 412 | 94 | 21.8% (17.9-25.9) | 1.21 | 0.86-1.69 | .27 |
| NRM . | N . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 396 | 73 | 19.9% (14.7-22.5) | Ref | ||
| Haplo-PTCy | 1039 | 203 | 12.1% (16.3-21.0) | 0.98 | 0.72-1.32 | .87 |
| MSD-PTCy | 396 | 73 | 18.5% (14.7-22.5) | Ref | ||
| Haplo: B-leader–matched | 627 | 109 | 16.5% (13.7-19.5) | 0.83 | 0.6-1.15 | .27 |
| Haplo: B-leader–mismatched | 412 | 94 | 21.8% (17.9-25.9) | 1.21 | 0.86-1.69 | .27 |
| Relapse∗ . | N . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 396 | 155 | 38.5% (33.5-43.5) | Ref | ||
| Haplo-PTCy | 1039 | 379 | 34.2% (31.3-37.2) | 0.87 | 0.7-1.08 | .20 |
| MSD-PTCy | 396 | 155 | 38.5% (33.5-43.5) | Ref | ||
| Haplo: DRB1–mismatched | 887 | 310 | 32.9% (29.7-36.0) | 0.82 | 0.66-1.02 | .07 |
| Haplo: DRB1–matched | 152 | 69 | 42.1% (34.0-49.9) | 1.14 | 0.85-1.54 | .39 |
| Relapse∗ . | N . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 396 | 155 | 38.5% (33.5-43.5) | Ref | ||
| Haplo-PTCy | 1039 | 379 | 34.2% (31.3-37.2) | 0.87 | 0.7-1.08 | .20 |
| MSD-PTCy | 396 | 155 | 38.5% (33.5-43.5) | Ref | ||
| Haplo: DRB1–mismatched | 887 | 310 | 32.9% (29.7-36.0) | 0.82 | 0.66-1.02 | .07 |
| Haplo: DRB1–matched | 152 | 69 | 42.1% (34.0-49.9) | 1.14 | 0.85-1.54 | .39 |
| Chronic GVHD . | N . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 399 | 131 | 33.6% (28.8-38.5) | Ref | ||
| Haplo-PTCy | 1035 | 333 | 31.7% (28.9-38.5) | 0.99 | 0.73-1.34 | .92 |
| Chronic GVHD . | N . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 399 | 131 | 33.6% (28.8-38.5) | Ref | ||
| Haplo-PTCy | 1035 | 333 | 31.7% (28.9-38.5) | 0.99 | 0.73-1.34 | .92 |
| Acute GVHD, grade 2-4† . | N . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 400 | 129 | 32.3% (27.7-36.9) | Ref | ||
| Haplo-PTCy | 1051 | 326 | 31.0% (28.3-33.9) | 0.95 | 0.71-1.27 | .74 |
| Acute GVHD, grade 2-4† . | N . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| MSD-PTCy | 400 | 129 | 32.3% (27.7-36.9) | Ref | ||
| Haplo-PTCy | 1051 | 326 | 31.0% (28.3-33.9) | 0.95 | 0.71-1.27 | .74 |
Full MVA tables are shown in supplemental Table 2.
Haplo, haploidentical; MVA, multivariate analysis; Ref, reference.
Stratified by disease stage.
Stratified by race.
Although donor type did not influence the risk of overall mortality, donor age emerged as a significant predictor. Each year increase in donor age was associated with 1% increase in overall mortality (hazard ratio [HR], 1.01; 95% confidence interval [CI], 1.001-1.014; P = .03; supplemental Table 2).
Younger haploidentical vs older MSD
With donor age affecting OS, we focused on outcomes for younger haploidentical donors compared with older MSDs (Table 1).
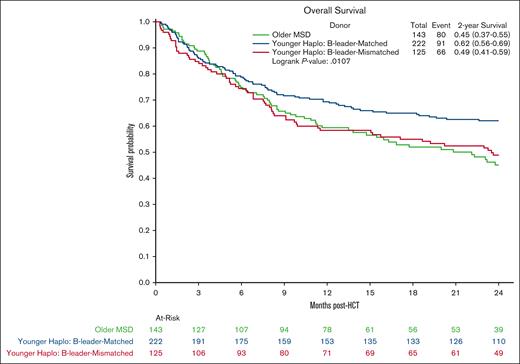
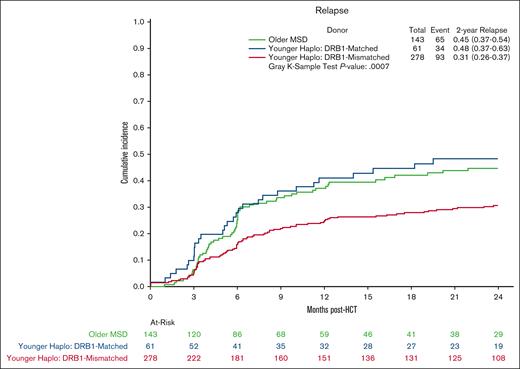
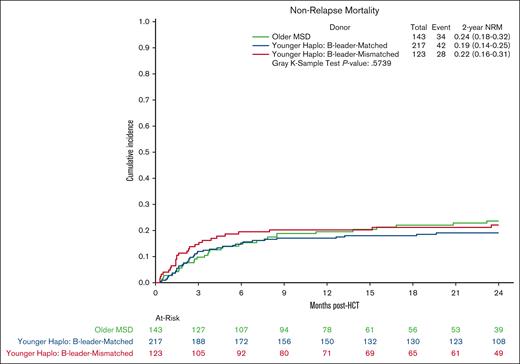
Multivariate analysis showed no significant differences in OS in the younger haploidentical group (HR, 0.76; 95% CI, 0.57-1.01; P = .06) compared with the older MSD. However, categorizing by HLA B-leader status revealed a significant OS benefit (HR, 0.65; 95% CI 0.48-0.90; P = .009) for the younger haploidentical B-leader matched but not B-leader mismatched group compared with the older MSD group (Table 3; supplemental Table 5; Kaplan Meier OS shown in Figure 2). A similar pattern emerged for relapse risk. The younger haploidentical group showed similar risk of relapse compared with older MSDs (HR, 0.71; 95% CI, 0.52-0.98; P = .04). However, further analysis based on HLA-DRB1 status identified a significantly lower relapse risk in the younger DRB1-mismatched but not DRB1-matched haploidentical group (HR, 0.63; 95% CI, 0.46-0.87; P = .004) compared with the older MSD group (cumulative incidence shown in Figure 3). No significant differences were noted in the risk of NRM (cumulative incidence shown in Figure 4), grade 2-4 acute GVHD, or chronic GVHD between the groups).
Multivariate analysis: younger haploidentical vs older MSD
| OS . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 143 | 80 | 45.1% (36.5-54.0) | Ref | ||
| Younger haplo-PTCy | 347 | 157 | 57.3 % (52.0-62.5) | 0.76 | 0.57-1.01 | .06 |
| Older MSD-PTCy | 143 | 80 | 45.1% (36.5-54.0) | Ref | ||
| Younger haplo: B-leader–matched | 222 | 91 | 62.1% (55.5-68.4) | 0.65 | 0.48-0.9 | .009 |
| Younger haplo: B-leader–mismatched | 125 | 66 | 48.9% (40.1-57.7) | 0.99 | 0.7-1.39 | .95 |
| OS . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 143 | 80 | 45.1% (36.5-54.0) | Ref | ||
| Younger haplo-PTCy | 347 | 157 | 57.3 % (52.0-62.5) | 0.76 | 0.57-1.01 | .06 |
| Older MSD-PTCy | 143 | 80 | 45.1% (36.5-54.0) | Ref | ||
| Younger haplo: B-leader–matched | 222 | 91 | 62.1% (55.5-68.4) | 0.65 | 0.48-0.9 | .009 |
| Younger haplo: B-leader–mismatched | 125 | 66 | 48.9% (40.1-57.7) | 0.99 | 0.7-1.39 | .95 |
| NRM . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 143 | 34 | 23.7% (16.9-31.1) | Ref | ||
| Younger haplo-PTCy | 340 | 70 | 20.1% (16-24.6) | 0.96 | 0.7-1.33 | .82 |
| Older MSD-PTCy | 143 | 34 | 23.7% (16.9-31.1) | Ref | ||
| Younger haplo: B-leader–matched | 217 | 42 | 19% (14.1-24.5) | 0.87 | 0.61-1.23 | .43 |
| Younger haplo: B-leader–mismatched | 123 | 28 | 22.1% (15.2-29.8) | 1.12 | 0.78-1.6 | .55 |
| NRM . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 143 | 34 | 23.7% (16.9-31.1) | Ref | ||
| Younger haplo-PTCy | 340 | 70 | 20.1% (16-24.6) | 0.96 | 0.7-1.33 | .82 |
| Older MSD-PTCy | 143 | 34 | 23.7% (16.9-31.1) | Ref | ||
| Younger haplo: B-leader–matched | 217 | 42 | 19% (14.1-24.5) | 0.87 | 0.61-1.23 | .43 |
| Younger haplo: B-leader–mismatched | 123 | 28 | 22.1% (15.2-29.8) | 1.12 | 0.78-1.6 | .55 |
| Relapse∗ . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 143 | 65 | 44.7% (36.1-52.9) | Ref | ||
| Younger haplo-PTCy | 340 | 127 | 33.7% (28.7-38.8) | 0.71 | 0.52-0.98 | .04 |
| Older MSD-PTCy | 143 | 65 | 44.7% (36.1-52.9) | Ref | ||
| Younger haplo: DRB1-mismatched | 278 | 93 | 30.6% (25.2-36.1) | 0.63 | 0.46-0.87 | .004 |
| Younger haplo: DRB1-matched | 61 | 34 | 48.3% (34.9-60.4) | 1.03 | 0.68-1.57 | .88 |
| Relapse∗ . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 143 | 65 | 44.7% (36.1-52.9) | Ref | ||
| Younger haplo-PTCy | 340 | 127 | 33.7% (28.7-38.8) | 0.71 | 0.52-0.98 | .04 |
| Older MSD-PTCy | 143 | 65 | 44.7% (36.1-52.9) | Ref | ||
| Younger haplo: DRB1-mismatched | 278 | 93 | 30.6% (25.2-36.1) | 0.63 | 0.46-0.87 | .004 |
| Younger haplo: DRB1-matched | 61 | 34 | 48.3% (34.9-60.4) | 1.03 | 0.68-1.57 | .88 |
| Chronic GVHD . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 142 | 47 | 33.3% (25.4-41.3) | Ref | ||
| Younger haplo-PTCy | 337 | 91 | 26.5% (21.9-31.4) | 0.78 | 0.49-1.25 | .31 |
| Chronic GVHD . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 142 | 47 | 33.3% (25.4-41.3) | Ref | ||
| Younger haplo-PTCy | 337 | 91 | 26.5% (21.9-31.4) | 0.78 | 0.49-1.25 | .31 |
| Acute GVHD, grade 2-4† . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 143 | 44 | 30.8% (23.4-38.5) | Ref | ||
| Younger haplo-PTCy | 347 | 97 | 27.9% (23.4-32.8) | 0.90 | 0.58-1.39 | .64 |
| Acute GVHD, grade 2-4† . | n . | Events . | Point estimates . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| Older MSD-PTCy | 143 | 44 | 30.8% (23.4-38.5) | Ref | ||
| Younger haplo-PTCy | 347 | 97 | 27.9% (23.4-32.8) | 0.90 | 0.58-1.39 | .64 |
Haplo, haploidentical; Ref, reference.
Stratified by disease stage.
Stratified by race.
Overall survival after MSD-PTCy HCT vs haploidentical (by B-leader match status). Kaplan Meier probability of OS comparing older HLA-MSD (green), younger haploidentical B-leader–matched donor (blue), and younger haploidentical B-leader–mismatched donor (red).
Overall survival after MSD-PTCy HCT vs haploidentical (by B-leader match status). Kaplan Meier probability of OS comparing older HLA-MSD (green), younger haploidentical B-leader–matched donor (blue), and younger haploidentical B-leader–mismatched donor (red).
Relapse after MSD-PTCy HCT vs haploidentical (by DRB1 match status). Cumulative incidence curves for relapse comparing older HLA-MSD (green), younger haploidentical DRB1-matched donor (blue), and younger haploidentical DRB1-mismatched donor (red).
Relapse after MSD-PTCy HCT vs haploidentical (by DRB1 match status). Cumulative incidence curves for relapse comparing older HLA-MSD (green), younger haploidentical DRB1-matched donor (blue), and younger haploidentical DRB1-mismatched donor (red).
Cumulative incidence curves for NRM comparing older HLA-MSD (green), younger haploidentical B-leader–matched donor (blue), and younger haploidentical B-leader–mismatched donor (red).
Cumulative incidence curves for NRM comparing older HLA-MSD (green), younger haploidentical B-leader–matched donor (blue), and younger haploidentical B-leader–mismatched donor (red).
Discussion
Our analysis yielded some key findings. First, haploidentical-PTCy demonstrated outcomes comparable with MSD-PTCy. Second, younger haploidentical donors, with B-leader matching were associated with improved OS and, or DRB1-mismatching was linked to reduced relapse risk, suggesting those 2 donor categories may potentially be preferable alternatives to older MSDs. These findings emphasize the critical role of considering HLA factors in comparisons between these donor types. Disregarding these factors in analyses may mask potential differences in transplant outcomes.
Younger haploidentical donor age has also been reported as a significant predictor of outcomes in other studies.3,16-20 The potential survival benefit observed with younger haploidentical donors in our study appears to be primarily driven by a reduction in relapse risk, rather than a decrease in NRM. This aligns with previous studies reporting a higher relapse risk associated with older MSDs compared with younger HLA-matched donors.8-10 Because relapse is the leading cause of death after HCT,2 achieving a reduction in relapse risk is expected to be 1 of the most impactful strategies for improving transplant outcomes.
Our limited sample size prevented a more nuanced exploration of donor selection based on additional factors. This included DQB1-matching and nonpermissive DPB1 mismatching in the haploidentical donors, previously linked to improved outcomes.3 Regrettably, the small size of the nonpermissive DPB1 mismatched subgroup (7%) and missing data (53%) precluded its use for categorization. Similarly, DQB1 matching was omitted because of sample size limitations and the high likelihood (>85%) of synchronous matching/mismatching at both DRB1 and DQB1 loci. Future studies with larger cohorts should incorporate these factors for a more comprehensive analysis. Furthermore, the lack of data on donor-specific antibodies, female donor parity, ABO matching, molecular disease classification, and pre-HCT measurable residual disease status limited the generalizability of our findings. Finally, the study only included patients who received transplantation up to 2017. The impact of advancements in supportive care since then remains unknown. Validation with larger, more recent data sets is necessary.
Conclusions
In conclusion, our study suggests that haploidentical-HCT and MSD-HCT with PTCy prophylaxis may offer comparable outcomes for patients with acute leukemia or MDS. Furthermore, among older patients (≥50 years), a younger haploidentical donor (<35 years) with either B-leader match (improved OS) and/or DRB1-mismatch (reduced relapse) might be preferable to an older MSD (≥50 years). These hypothesis-generating findings warrant validation in larger studies.
Acknowledgments
The authors acknowledge that European Union patient-level data are redacted. The authors are grateful to the Center for International Blood and Marrow Transplant Research (CIBMTR) staff for providing the data sets publicly.
This dataset was collected by the Center for International Blood and Marrow Transplant Research which is supported primarily by the Public Health Service (U24CA076518) from the National Cancer Institute; the National Heart, Lung, and Blood Institute; the National Institute of Allergy and Infectious Diseases; 75R60222C00011 from the Health Resources and Services Administration; N00014-23-1-2057 and N00014-24-1-2057 from the Office of Naval Research; NMDP; and the Medical College of Wisconsin.
Authorship
Contribution: R.S.M. conceptualized the study, performed the statistical analysis, interpreted data, wrote the manuscript, and had the final responsibility to submit for publication; J.R., P.K., U.P., R.E.C., K.R., and E.J.S. reviewed and interpreted the data, reviewed the manuscript, and provided critical feedback; R.S.M. had full access to the raw data, which are publicly available; and all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rohtesh S. Mehta, Clinical Research Division, University of Washington School of Medicine, Fred Hutchinson Cancer Center, 1100 Fairview Ave N, Mail Stop M2-B235, Seattle, WA 98109; email: rmehta@fredhutch.org.
References
Author notes
Data are publicly available and accessible at https://cibmtr.org/CIBMTR/Resources/Publicly-Available-Datasets.
The full-text version of this article contains a data supplement.