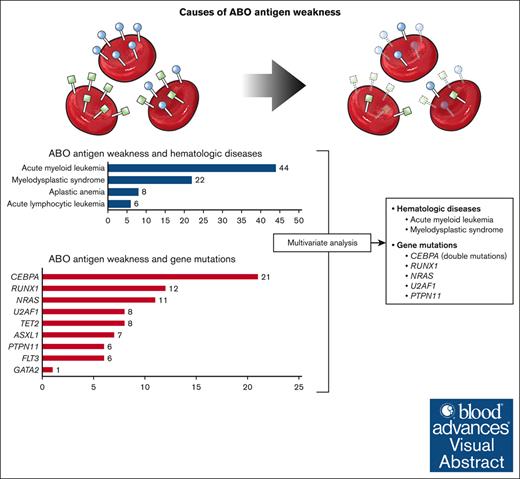
ABO antigen weakness is associated with mutations of RUNX1, CEBPA, NRAS, U2AF1, and PTPN11.
CEBPA double mutations were closely related to ABO antigen weakness, whereas single mutation showed no such association.
Visual Abstract
ABO antigen weakness is rarely observed in ABO typing for transfusion. Hematologic diseases and associated gene mutations have been suggested as potential causes of this phenomenon, yet the precise etiology has not been elucidated. Through ABO typing and genetic analysis data conducted over 7 years, we have reconfirmed the association between ABO antigen weakness and hematologic diseases, especially acute myeloid leukemia (odds ratio [OR], 2.55; 95% confidence interval [CI], 1.12-5.83) and myelodysplastic syndrome (OR, 6.94; 95% CI, 2.86-16.83), and discovered previously unidentified candidate genes, CEBPA (OR, 43.70; 95% CI, 18.12-105.40), NRAS (OR, 3.37; 95% CI, 1.46-7.79), U2AF1 (OR, 8.12; 95% CI, 2.86-23.03), and PTPN11 (OR, 4.52; 95% CI, 1.51-13.50), seemingly associated with this phenomenon. Among these, CEBPA double mutations displayed a significant association, with ABO antigen weakness being observed in 20 of the 25 individuals (80.0%) possessing these mutations. From this study, new factors associated with ABO antigen weakness have been identified.
Introduction
Weakened, or loss of, ABO antigens have been reported in patients with various clinical conditions.1,2 Weak ABO antigens may be attributed to ABO subgroups, such as A3, cis-AB, and so on, which were reported to comprise 0.05% to 0.13% of the Korean population.3,4 Besides those ABO subgroups, pregnancy1 and malignancies5,6 also cause alteration of ABO antigen expression. Especially, among malignancies, hematologic malignancies have been known for their association with ABO antigen weakness.1,2,7-9 Efforts have been made to identify factors associated with ABO antigen weakness in patients with hematologic malignancies. Mutations of genes involved in hematopoiesis, which were frequently detected in those patients, have been suspected as causes of the phenomenon.10,11
ABO antigen weakness causes discrepancies of pretransfusion ABO testing, which may cause confusion in transfusion practice. Understanding the phenomenon can guide practitioners to make appropriate transfusion decisions for patients with ABO antigen weakness. In addition, identification of factors such as specific gene mutations may lead to revealing the mechanism of ABO antigen weakness and further applications.
In our review of ABO typing results over a 7-year period, we identified ABO antigen weakness in 511 patients. Among these, 145 patients had malignancies, of which 103 were diagnosed with hematologic diseases. Genetic analysis of patients with hematologic diseases revealed not only the previously reported gene RUNX110,11 but also previously unidentified genes, CEBPA, NRAS, U2AF1, and PTPN11, all potentially associated with this phenomenon.
Methods
Study population and data collection
From January 2017 to June 2023, ABO typing was performed for 361 900 patients. Among them, 511 patients with weakened or loss of ABO antigens were included in the study. Diagnosis, age, gender, and past history of malignancy and hematologic diseases of each individual were collected. For patients with hematologic diseases who underwent genetic tests for the diseases, the results were also collected and included in the analysis. This study was reviewed and approved by the Institutional Review Board (IRB) of Yonsei University Health System, Severance Hospital in Seoul, Korea (IRB approval number 4-2023-0606). The informed consent requirement for the inclusion of this study was waived due to the retrospective nature of this study.
ABO typing
ABO typing was performed by automated instrument using IH-1000 (Bio-Rad Laboratories, Hercules, CA), VISION MAX and AutoVue Innova (Ortho-Clinical Diagnostics, Raritan, NJ). In addition, manual ABO typing was conducted by tube method as our laboratory’s standard operating procedure. Sihdia Anti-A, Anti-B Blood Grouping Serum (Shinyang Chemical, Seoul, Korea) were used for front typing. Approximately 2% to 5% of the patient red blood cells (RBCs) were mixed with Anti-A or Anti-B antisera and centrifuged at 3400 rpm for 15 seconds at room temperature. In addition, Anti-A1 lectin (Lorne Laboratories LTD, Reading, Berkshire, UK) was also used for front typing for accurate evaluation of ABO group. For back typing, 2 drops of patient’s serum were mixed with A1 or B RBC (iMR.A1-B; MIRR. SciTech Corp, Seoul, Korea). Additional steps during the ABO front typing process include washing the patient's RBCs before front typing and implementing an extra incubation time during front typing when necessary. After reaction, the degree of hemagglutination was scored as follows: negative, ±, 1+, 2+, 3+, or 4+. If observed, the presence of mixed field agglutination was also noted. ABO antigen weakness was defined when the hemagglutination was less than 4+ and/or mixed field agglutination was detected, either from automated instrument or manual typing.
ABO genotyping
Genomic DNA (gDNA) was extracted from bone marrow aspirate using the QIAamp Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s guidelines. All coding exons and nearby splicing sites, proximal promoter, CCAAT-binding factor/NF-Y binding site, and +5.8-kb site of ABO were sequenced using a 3730 DNA Analyzer with BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA).
Variant analysis using next-generation sequencing data for hematologic diseases
To investigate the association between ABO antigen weakness and specific gene abnormalities, mutation information was retrospectively collected from the reports of hematologic next-generation sequencing performed at Severance Hospital for the diagnosis of hematologic diseases. The next-generation sequencing for the diagnosis of hematologic diseases was conducted by extracting gDNA from bone marrow aspirate. Extracted gDNA was quantified using the Qubit BR dsDNA kit (Invitrogen, Carlsbad, CA). For targeted next-generation sequencing for the diagnosis of hematologic diseases, libraries were prepared, and targets were captured using a custom panel (Dxome, Seoul, Korea) consisting of genes listed in supplemental Table 1. The prepared libraries were sequenced on the NextSeq 550Dx instrument (Illumina, San Diego, CA). The sequencing data were analyzed using the DxSeq Analyzer (Dxome).
Detected variants were classified into 4 tiers based on their level of clinical significance in cancer diagnosis, prognosis, and/or therapeutics following the standards and guidelines established by the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists,12 from tier 1 to tier 4. Only tier 1 and 2 variants were included in the final analysis.
Statistical analysis
Fisher exact test was performed to examine the association between the presence of certain gene mutations and ABO antigen weakness. To figure out the factors related to ABO antigen weakness, logistic regression was conducted for patients who underwent genetic tests for hematologic diseases. Diagnosis and genes with mutations were included in the analysis. All statistical analyses were conducted using SPSS version 26.0 (IBM Corp, Armonk, NY), with P values of < .05 considered statistically significant.
Results
ABO antigen weakness identified from patients with various clinical conditions
ABO antigen weakness was identified from 511 individuals among a total of 361 900 individuals tested (0.14%). The demographics of the participants whose ABO antigen weakness was detected have been compiled in Table 1. Mean age of individuals with weakened ABO antigen was 50.4 years. The phenomenon of ABO antigen weakness showed a similar occurrence rate between males (n = 250 [48.9%]) and females (n = 261 [51.1%]). ABO antigen weakness was most frequently observed in patients with malignancies (n = 145 [28.4%]), followed by those with hematologic diseases (n = 103 [28.4%]). This phenomenon was also observed in pregnant women (n = 18 [3.5%]) and individuals without any underlying health conditions (n = 47 [9.2%]).
Demographics of patients with ABO antigen weakness
| . | Total (n = 511) . | A weak (n = 172) . | B weak (n = 309) . | Both (n = 30) . |
|---|---|---|---|---|
| Age, mean, y | 50.4 | 50.7 | 50.5 | 47.7 |
| Sex | ||||
| Male | 250 (48.9%) | 90 (52.3%) | 144 (46.4%) | 16 (53.3%) |
| Female | 261 (51.1%) | 82 (47.7%) | 165 (53.4%) | 14 (46.7%) |
| Clinical status | ||||
| Malignancy | 145 (28.4%) | 45 (26.2%) | 93 (30.1%) | 7 (23.3%) |
| Chemotherapy | 82 (16.0%) | 31 (18.0%) | 41 (13.3%) | 10 (33.3%) |
| Hematologic diseases | 103 (20.2%) | 55 (32.0%) | 32 (10.4%) | 16 (53.3%) |
| Pregnancy | 18 (3.5%) | 4 (2.3%) | 13 (4.2%) | 1 (3.3%) |
| Healthy | 47 (9.2%) | 12 (7.0%) | 33 (10.7%) | 2 (6.7%) |
| . | Total (n = 511) . | A weak (n = 172) . | B weak (n = 309) . | Both (n = 30) . |
|---|---|---|---|---|
| Age, mean, y | 50.4 | 50.7 | 50.5 | 47.7 |
| Sex | ||||
| Male | 250 (48.9%) | 90 (52.3%) | 144 (46.4%) | 16 (53.3%) |
| Female | 261 (51.1%) | 82 (47.7%) | 165 (53.4%) | 14 (46.7%) |
| Clinical status | ||||
| Malignancy | 145 (28.4%) | 45 (26.2%) | 93 (30.1%) | 7 (23.3%) |
| Chemotherapy | 82 (16.0%) | 31 (18.0%) | 41 (13.3%) | 10 (33.3%) |
| Hematologic diseases | 103 (20.2%) | 55 (32.0%) | 32 (10.4%) | 16 (53.3%) |
| Pregnancy | 18 (3.5%) | 4 (2.3%) | 13 (4.2%) | 1 (3.3%) |
| Healthy | 47 (9.2%) | 12 (7.0%) | 33 (10.7%) | 2 (6.7%) |
When looking at the frequency of weakened antigens for each ABO group, A antigen weakness was detected in 172 individuals (33.7%), whereas weakened B antigen was identified in 309 individuals (60.5%). Weakness of both A and B antigens was observed in 30 individuals (5.9%). The distribution of each antigen weakness in terms of age and sex was similar, with B antigen weakness being the most commonly observed, followed by A antigen weakness. When looking at the frequency of antigen weakness by clinical situations, the distribution was also similar, except in the case of hematologic disease, in which A antigen weakness was most frequently observed (55/172 [32.0%]).
In addition, we further reviewed patients with malignancies. ABO antigen weakness was most frequently observed in patients with stomach cancer (20/145 [13.8%]), followed by lung (n = 18 [12.4%]), thyroid (n = 18 [12.4%]), breast (n = 13 [9.0%]), prostate (n = 9 [6.2%]), and ovary (n = 7 [4.8%]) cancers. Among the patients with malignancies and ABO antigen weakness, 43 individuals (29.7%) had undergone chemotherapy treatment within 1 year before ABO antigen weakness, whereas 7 (4.8%) had received radiotherapy (supplemental Table 2).
ABO antigen weakness identified from patients with hematologic diseases
Among individuals with ABO antigen weakness, patients with hematologic disease were further investigated (Table 2). Among them, acute myeloid leukemia (AML) was the most common (44/103 [42.7%]), followed by myelodysplastic syndrome (MDS) (n = 22 [21.4%]). Genetic analysis was conducted in 64 patients. Mutations of CEBPA were the most common genetic aberration (21/64 [32.8%]). Mutations of RUNX1 (n = 12 [18.8%]) and NRAS (n = 11 [17.2%]) were also frequently identified from patients with weakened ABO antigens. There was only 1 case with a pathogenic variant of GATA2. Other frequently detected genes with pathogenic variants were TET2, U2AF1, ASXL1, FLT3, and PTPN11.
Demographics, diagnosis, and gene mutations in patients with ABO antigen weakness and hematologic diseases
| . | Total (n = 103) . | A weak (n = 55) . | B weak (n = 32) . | Both (n = 16) . |
|---|---|---|---|---|
| Sex | ||||
| Male | 57 (55.3) | 30 (54.5) | 19 (59.4) | 8 (50.0) |
| Female | 46 (44.7) | 25 (45.5) | 13 (40.6) | 8 (50.0) |
| Age, mean, y | 51.4 | 51.8 | 51.1 | 50.9 |
| Hematologic diseases | ||||
| AML | 44 (42.7) | 23 (41.8) | 12 (37.5) | 9 (56.3) |
| ALL | 6 (5.8) | 5 (9.1) | 0 (0.0) | 1 (6.3) |
| MDS | 22 (21.4) | 12 (21.8) | 10 (31.3) | 0 (0.0) |
| AA | 8 (7.8) | 5 (9.1) | 1 (3.1) | 2 (12.5) |
| Genetic tested∗ | 64 | 35 | 19 | 10 |
| RUNX1 | 12 (18.8) | 8 (22.9) | 2 (10.5) | 2 (20.0) |
| GATA2 | 1 (1.6) | 0 (0.0) | 0 (0.0) | 1 (10.0) |
| CEBPA | 21 (32.8) | 12 (34.3) | 5 (26.3) | 4 (40.0) |
| NRAS | 11 (17.2) | 4 (11.4) | 4 (21.1) | 3 (30.0) |
| TET2 | 8 (12.5) | 3 (8.6) | 3 (15.8) | 2 (20.0) |
| U2AF1 | 8 (12.5) | 3 (8.6) | 5 (26.3) | 0 (0.0) |
| ASXL1 | 7 (10.9) | 6 (17.1) | 0 (0.0) | 1 (10.0) |
| FLT3 | 6 (9.4) | 4 (11.4) | 1 (5.3) | 1 (10.0) |
| PTPN11 | 6 (9.4) | 4 (11.4) | 1 (5.3) | 1 (10.0) |
| . | Total (n = 103) . | A weak (n = 55) . | B weak (n = 32) . | Both (n = 16) . |
|---|---|---|---|---|
| Sex | ||||
| Male | 57 (55.3) | 30 (54.5) | 19 (59.4) | 8 (50.0) |
| Female | 46 (44.7) | 25 (45.5) | 13 (40.6) | 8 (50.0) |
| Age, mean, y | 51.4 | 51.8 | 51.1 | 50.9 |
| Hematologic diseases | ||||
| AML | 44 (42.7) | 23 (41.8) | 12 (37.5) | 9 (56.3) |
| ALL | 6 (5.8) | 5 (9.1) | 0 (0.0) | 1 (6.3) |
| MDS | 22 (21.4) | 12 (21.8) | 10 (31.3) | 0 (0.0) |
| AA | 8 (7.8) | 5 (9.1) | 1 (3.1) | 2 (12.5) |
| Genetic tested∗ | 64 | 35 | 19 | 10 |
| RUNX1 | 12 (18.8) | 8 (22.9) | 2 (10.5) | 2 (20.0) |
| GATA2 | 1 (1.6) | 0 (0.0) | 0 (0.0) | 1 (10.0) |
| CEBPA | 21 (32.8) | 12 (34.3) | 5 (26.3) | 4 (40.0) |
| NRAS | 11 (17.2) | 4 (11.4) | 4 (21.1) | 3 (30.0) |
| TET2 | 8 (12.5) | 3 (8.6) | 3 (15.8) | 2 (20.0) |
| U2AF1 | 8 (12.5) | 3 (8.6) | 5 (26.3) | 0 (0.0) |
| ASXL1 | 7 (10.9) | 6 (17.1) | 0 (0.0) | 1 (10.0) |
| FLT3 | 6 (9.4) | 4 (11.4) | 1 (5.3) | 1 (10.0) |
| PTPN11 | 6 (9.4) | 4 (11.4) | 1 (5.3) | 1 (10.0) |
AA, aplastic anemia; ALL, acute lymphocytic leukemia.
Numbers and proportions of patients with each gene mutation among tested patients were described. Only tier 1, 2 variants of each gene were counted.
Because ABO antigen weakness can be observed in patients with ABO subgroups, ABO genotyping was conducted on a subset of patients for whom samples were available (31 of 64 patients who underwent genetic tests). Except for 1 patient who exhibited a reduced A allele due to a copy number variation on chromosome 9, the other 30 patients did not show any variants associated with ABO subgroups (supplemental Table 3).
Genes associated with ABO antigen weakness
For genes in which mutations were frequently detected in patients with ABO antigen weakness, their association with the phenomenon was further investigated (Table 3). For this, additional analyses were performed on patients who received genetic tests for the diagnosis of hematologic diseases between 2017 and 2023, regardless of the presence of ABO antigen weakness. Among these, a total of 1445 patients, excluding those of blood type O and those who did not undergo ABO testing, were included in the analysis. Among 61 patients with RUNX1 mutations, ABO antigen weakness was detected from 12 (19.7%) and significantly correlated with the presence of RUNX1 mutations (P < .001). Mutations of CEBPA were identified in 39 patients, with 21 patients with weakened ABO antigens (53.8%). The presence of CEBPA mutations were also significantly associated with the phenomenon (P < .001). Antigen weakness was detected in 11 patients with NRAS mutations (11/103 [10.7%]), and these mutations were significantly associated with the phenomenon of antigen weakening (P = .004). Other genes, such as TET2, U2AF1, ASXL1, and PTPN11, showed an association with the phenomenon, whereas GATA2 (P = .272) and FLT3 (P = .444) did not exhibit any significant correlation.
Association between ABO antigen weakness and each gene with mutations
| . | ABO antigen weakness . | P value . | ||
|---|---|---|---|---|
| RUNX1 mutation | Weakness− | Weakness+ | Sum | < .001 |
| Mutation− | 1332 | 52 | 1384 | |
| Mutation+ | 49 | 12 | 61 | |
| Sum | 1381 | 64 | 1445 | |
| GATA2 mutation | Weakness− | Weakness+ | Sum | .272 |
| Mutation− | 1375 | 63 | 1438 | |
| Mutation+ | 6 | 1 | 7 | |
| Sum | 1381 | 64 | 1445 | |
| CEBPA mutation | Weakness− | Weakness+ | Sum | < .001 |
| Mutation− | 1363 | 43 | 1406 | |
| Mutation+ | 18 | 21 | 39 | |
| Sum | 1381 | 64 | 1445 | |
| NRAS mutation | Weakness− | Weakness+ | Sum | .004 |
| Mutation− | 1289 | 53 | 1342 | |
| Mutation+ | 92 | 11 | 103 | |
| Sum | 1381 | 64 | 1445 | |
| TET2 mutation | Weakness− | Weakness+ | Sum | .043 |
| Mutation− | 1286 | 55 | 1341 | |
| Mutation+ | 95 | 9 | 104 | |
| Sum | 1381 | 64 | 1445 | |
| U2AF1 mutation | Weakness− | Weakness+ | Sum | < .001 |
| Mutation− | 1364 | 56 | 1420 | |
| Mutation+ | 17 | 8 | 25 | |
| Sum | 1381 | 64 | 1445 | |
| ASXL1 mutation | Weakness− | Weakness+ | Sum | .022 |
| Mutation− | 1323 | 57 | 1380 | |
| Mutation+ | 58 | 7 | 65 | |
| Sum | 1381 | 64 | 1445 | |
| FLT3 mutation | Weakness− | Weakness+ | Sum | .444 |
| Mutation− | 1287 | 58 | 1345 | |
| Mutation+ | 94 | 6 | 100 | |
| Sum | 1381 | 64 | 1445 | |
| PTPN11 mutation | Weakness− | Weakness+ | Sum | .004 |
| Mutation− | 1351 | 58 | 1409 | |
| Mutation+ | 30 | 6 | 36 | |
| Sum | 1381 | 64 | 1445 | |
| . | ABO antigen weakness . | P value . | ||
|---|---|---|---|---|
| RUNX1 mutation | Weakness− | Weakness+ | Sum | < .001 |
| Mutation− | 1332 | 52 | 1384 | |
| Mutation+ | 49 | 12 | 61 | |
| Sum | 1381 | 64 | 1445 | |
| GATA2 mutation | Weakness− | Weakness+ | Sum | .272 |
| Mutation− | 1375 | 63 | 1438 | |
| Mutation+ | 6 | 1 | 7 | |
| Sum | 1381 | 64 | 1445 | |
| CEBPA mutation | Weakness− | Weakness+ | Sum | < .001 |
| Mutation− | 1363 | 43 | 1406 | |
| Mutation+ | 18 | 21 | 39 | |
| Sum | 1381 | 64 | 1445 | |
| NRAS mutation | Weakness− | Weakness+ | Sum | .004 |
| Mutation− | 1289 | 53 | 1342 | |
| Mutation+ | 92 | 11 | 103 | |
| Sum | 1381 | 64 | 1445 | |
| TET2 mutation | Weakness− | Weakness+ | Sum | .043 |
| Mutation− | 1286 | 55 | 1341 | |
| Mutation+ | 95 | 9 | 104 | |
| Sum | 1381 | 64 | 1445 | |
| U2AF1 mutation | Weakness− | Weakness+ | Sum | < .001 |
| Mutation− | 1364 | 56 | 1420 | |
| Mutation+ | 17 | 8 | 25 | |
| Sum | 1381 | 64 | 1445 | |
| ASXL1 mutation | Weakness− | Weakness+ | Sum | .022 |
| Mutation− | 1323 | 57 | 1380 | |
| Mutation+ | 58 | 7 | 65 | |
| Sum | 1381 | 64 | 1445 | |
| FLT3 mutation | Weakness− | Weakness+ | Sum | .444 |
| Mutation− | 1287 | 58 | 1345 | |
| Mutation+ | 94 | 6 | 100 | |
| Sum | 1381 | 64 | 1445 | |
| PTPN11 mutation | Weakness− | Weakness+ | Sum | .004 |
| Mutation− | 1351 | 58 | 1409 | |
| Mutation+ | 30 | 6 | 36 | |
| Sum | 1381 | 64 | 1445 | |
Given the presence of patients with multiple mutations and the potential association between the existence of hematologic disease and ABO antigen weakness, we conducted an analysis adjusted for both the presence or absence of disease and other gene mutations in patients who had undergone genetic testing as part of their hematologic disease diagnosis (Table 4). Alongside the presence of hematologic diseases, we also analyzed the occurrence of each gene mutation to determine their relationship with ABO antigen weakness. As known previously, AML (odds ratio, 2.55; 95% confidence interval, 1.12-5.83; P = .026) and MDS (odds ratio, 6.94; 95% confidence interval, 2.86-16.83; P < .001) were significantly associated with the phenomenon. Lymphoid malignancies, acute lymphocytic leukemia and lymphoma, as well as aplastic anemia were not associated with the phenomenon. Among genes, RUNX1, CEBPA, NRAS, U2AF1, and PTPN11 were shown to be associated with ABO antigen weakness.
Univariable and multivariable analysis of risk factors, hematologic diseases, and related gene mutations for ABO antigen weakness
| Factors . | Univariable . | Multivariable . | ||
|---|---|---|---|---|
| Odds ratio (95% CI) . | P value . | Odds ratio (95% CI) . | P value . | |
| AML | 4.16 (2.50-6.92) | < .001 | 2.55 (1.12-5.83) | .026 |
| MDS | 3.60 (2.00-6.46) | < .001 | 6.94 (2.86-16.83) | < .001 |
| ALL | 0.60 (0.24-1.53) | .286 | ||
| AA | 0.80 (0.11-5.95) | .824 | ||
| RUNX1 | 6.27 (3.15-12.50) | < .001 | 4.25 (1.77-10.21) | .001 |
| GATA2 | 3.64 (0.43-30.67) | .235 | ||
| CEBPA | 36.98 (18.38-74.39) | < .001 | 43.70 (18.12-105.40) | < .001 |
| NRAS | 2.91 (1.47-5.76) | .002 | 3.37 (1.46-7.79) | .005 |
| TET2 | 2.22 (1.06-4.62) | .034 | 0.85 (0.35-2.11) | .730 |
| U2AF1 | 11.46 (4.75-27.68) | < .001 | 8.12 (2.86-23.03) | < .001 |
| ASXL1 | 2.80 (1.22-6.41) | .015 | 0.69 (0.25-1.91) | .477 |
| FLT3 | 1.42 (0.60-3.37) | .431 | ||
| PTPN11 | 4.66 (1.87-11.63) | .001 | 4.52 (1.51-13.50) | .007 |
| Factors . | Univariable . | Multivariable . | ||
|---|---|---|---|---|
| Odds ratio (95% CI) . | P value . | Odds ratio (95% CI) . | P value . | |
| AML | 4.16 (2.50-6.92) | < .001 | 2.55 (1.12-5.83) | .026 |
| MDS | 3.60 (2.00-6.46) | < .001 | 6.94 (2.86-16.83) | < .001 |
| ALL | 0.60 (0.24-1.53) | .286 | ||
| AA | 0.80 (0.11-5.95) | .824 | ||
| RUNX1 | 6.27 (3.15-12.50) | < .001 | 4.25 (1.77-10.21) | .001 |
| GATA2 | 3.64 (0.43-30.67) | .235 | ||
| CEBPA | 36.98 (18.38-74.39) | < .001 | 43.70 (18.12-105.40) | < .001 |
| NRAS | 2.91 (1.47-5.76) | .002 | 3.37 (1.46-7.79) | .005 |
| TET2 | 2.22 (1.06-4.62) | .034 | 0.85 (0.35-2.11) | .730 |
| U2AF1 | 11.46 (4.75-27.68) | < .001 | 8.12 (2.86-23.03) | < .001 |
| ASXL1 | 2.80 (1.22-6.41) | .015 | 0.69 (0.25-1.91) | .477 |
| FLT3 | 1.42 (0.60-3.37) | .431 | ||
| PTPN11 | 4.66 (1.87-11.63) | .001 | 4.52 (1.51-13.50) | .007 |
AA, aplastic anemia; ALL, acute lymphocytic leukemia; CI, confidence interval.
Gene mutations and ABO antigen weakness
Although several genes were proven to be associated with the occurrence of ABO antigen weakness, the phenomenon was not observed in all patients with pathogenic variants of those genes. Consequently, to ascertain whether specific domains or variants of each gene were linked to the phenomenon, further investigation was conducted. The variants of each gene were listed according to the presence of ABO antigen weakness (supplemental Table 4). Although the association of RUNX1, NRAS, U2AF1, and PTPN11 with ABO antigen weakness has been proven, certain variant type or region that were related with the phenomenon could not be identified. Especially, almost all variants of NRAS, U2AF1, and PTPN11 detected from patients with ABO antigen weakness were also present in patients without ABO antigen weakness.
CEBPA was suspected to be strongly correlated with ABO antigen weakness, in which the phenomenon was identified from about half of the patients with pathogenic CEBPA variants. It has been known that patients with hematologic malignancies carrying CEBPA pathogenic variants can have either 1 (CEBPA single mutation, CEBPAsm) or 2 CEBPA mutations (CEBPA double mutations, CEBPAdm), and among 39 patients with CEBPA mutations in our study population, there were 25 patients with CEBPAdm and 14 patients with CEBPAsm. When analyzed separately, there were striking difference in association with ABO antigen weakness between CEBPAdm and CEBPAsm. While ABO antigen weakness was observed in 20 of 25 patients (80%) with CEBPAdm, the phenomenon was observed in only 1 of 14 patients (7.1%) with CEBPAsm. In addition, CEBPAsm was shown to not be associated with ABO antigen weakness (P = .471; Table 5). Similar to the other genes, when examining the mutations of CEBPA according to the presence or absence of antigen weakness, no evidence could be found that a specific genetic variant or a specific gene location was associated with the antigen weakness phenomenon (supplemental Table 5). However, there is an unusual circumstance with a patient (patient 2 from supplemental Table 5). The ABO blood typing was conducted after chemotherapy had initiated and when leukemic blasts had already disappeared from the peripheral blood. This leaves a possibility that the patient's ABO antigens might have been weak at the time of the initial diagnosis. Two mutations of a patient (patient 4 in supplemental Table 5), a heterozygous whole gene deletion and a heterozygous frameshift variant, were detected, which suggests the presence of different clones that harbored each variant.
The association between ABO antigen weakness and CEBPA double (CEBPAdm) or single (CEBPAsm) mutation
| . | ABO antigen weakness . | P value . | ||
|---|---|---|---|---|
| CEBPAdm | Weakness− | Weakness+ | Sum | < .001 |
| Mutation− | 1376 | 44 | 1420 | |
| Mutation+ | 5 | 20 | 25 | |
| Sum | 1381 | 64 | 1445 | |
| CEBPAsm | Weakness− | Weakness+ | Sum | .471 |
| Mutation− | 1368 | 63 | 1431 | |
| Mutation+ | 13 | 1 | 14 | |
| Sum | 1381 | 64 | 1445 | |
| . | ABO antigen weakness . | P value . | ||
|---|---|---|---|---|
| CEBPAdm | Weakness− | Weakness+ | Sum | < .001 |
| Mutation− | 1376 | 44 | 1420 | |
| Mutation+ | 5 | 20 | 25 | |
| Sum | 1381 | 64 | 1445 | |
| CEBPAsm | Weakness− | Weakness+ | Sum | .471 |
| Mutation− | 1368 | 63 | 1431 | |
| Mutation+ | 13 | 1 | 14 | |
| Sum | 1381 | 64 | 1445 | |
Discussion
Weakened ABO antigens are occasionally encountered during ABO testing for blood transfusion. Several related factors have been identified, and there have been reports suggesting an association with diseases such as AML. However, most of these reports were sporadic case reports. In this study, factors associated with ABO antigen weakness were comprehensively analyzed, including a re-evaluation of previously known associated factors and the identification of additional potential associated factors.
ABO antigen weakness of patients with malignancies has been reported. However, it seems that malignancies only affect ABO expression in cancer tissues.5,6 There were reports of RBC ABO antigen weakness in patients with cancer, however, ABO antigen weakness in those patients were due to excessive ABO antigen that affect ABO typing results, rather than weakness of antigens on RBCs themselves.13,14 We sought to find whether specific type of cancer or history of anticancer therapy were related with decreased ABO antigen expression on RBC. Although ABO antigen weakness was found in patients with various types of cancers including thyroid, lung, stomach, and others, the proportion of each cancer type was similar to the incidence of each cancer type in the Korean population. Moreover, the association of chemotherapy and radiotherapy was not apparent. Based on the information above, we found it challenging to establish a clear relationship between malignancies and the ABO antigen weakness.
The ABO antigen weakness during pregnancy was also reported.1 In that previous study, all identified weakened ABO antigen was blood type A, and it was hypothesized that hormonal change during pregnancy might affect ABO antigen expression. In contrast, in our data, A antigen weakness was observed in only 5 of 18 cases (27.8%; 4 in A blood type and 1 in AB blood type) among pregnant individuals with antigen weakness. This proportion was similar to the rate of A antigen weakness cases among the overall healthy population with antigen weakness (14/47 [29.8%]). Therefore, based on the results of this study, we were unable to establish a significant association between pregnancy and antigen weakness.
The association between ABO antigen weakness and hematologic diseases is well established. This phenomenon has primarily been observed in patients with AML and MDS. However, the mechanism underlying ABO antigen weakness in these patients has not been fully understood. It has been suggested that genetic abnormalities in hematopoietic stem cells may be the cause of this phenomenon. Although newly developed genetic variants of the ABO gene could be suspected, ABO genotyping performed on patients with this phenomenon did not reveal any newly introduced genetic abnormalities in these individuals, which was similar to our data.15,16 In addition, other genes known to be involved in ABO gene expression were also investigated, and mutations of RUNX1 and GATA2 were suggested to be related with the reduction of erythrocyte A antigen expression.10,11RUNX1 encodes a Runt-related transcription factor, which plays a role in regulation of hematopoiesis.17RUNX1 is one of the frequently mutated genes in myeloid malignancies.18GATA2 encodes a transcription factor that have pivotal roles in hematopoiesis.19 Although the mechanism was not clarified, the presence of a GATA2 pathogenic variant in patients with AML with reduced A antigen expression suggests that GATA2 is implicated as a potential cause of antigenic attenuation.11 Although previous studies suggested the potential involvement of RUNX1 and GATA2 variants in weakening ABO antigen expression, the evidence for this association remains limited to a few cases and simple in vitro assays using cell lines. A recent study investigating the association of RUNX1 and GATA2 with this phenomenon among patients with AML found no significant correlation between these genes and ABO antigen weakness.20 To address the discrepancies observed in previous studies, this study aimed to identify genes associated with the phenomenon by analyzing genetic test results from patients. The results revealed a significant association between RUNX1 and ABO antigen weakness, whereas no association was identified with GATA2. Despite the confirmed association, there are numerous patients who have RUNX1 mutations but do not exhibit ABO antigen weakness. In addition, among patients with RUNX1 mutations, no specific characteristics associated with antigen weakening have been identified.
However, the analysis unveiled previously unrecognized genes potentially involved in this phenomenon. Among them, CEBPAdm showed strong association with the ABO antigen weakness. A transcription factor, CCAAT-enhancer binding protein α (C/EBPα), is encoded by CEBPA and involved in myeloid development.21 Mutated CEBPA is detected in 7.5% to 18% of patients with myeloid malignancies.22 Until now, there has been no reported association between the expression of ABO antigens or the relationship between ABO and CEBPA. However, it has been reported that there is a CEBPA binding site in the +5.8-kb region of the ABO, which is known to be related to the regulation of ABO gene expression, and a mutation in adjacent areas have been associated with ABO antigen weakness.23 From these observations, it can be postulated that CEBPA may play a role in modulating the ABO gene expression, potentially leading to the manifestation of antigenic weakness. In addition, the observation that only double mutations in CEBPA are associated with the antigen weakness, whereas single mutations are not, suggests that the presence of a functional CEBPA may be implicated in the expression of ABO antigens.
Interestingly, previous reports indicating a decrease in CEBPA expression in RUNX1-mutated AML24 present the possibility that the observed ABO antigen weakness associated with RUNX1 mutations may also be related to CEBPA. This underscores the need for further research to explore this potential connection.
Association of other genes, NRAS, PTPN11, and U2AF1, with ABO antigen weakness were also revealed from the analysis. NRAS, a member of RAS oncogene family, was one of the frequently mutated genes in AML.25 Another gene involved in RAS signaling pathway, PTPN11, is mutated in a small proportion of patients with AML.26 Mutations of U2AF1, which encodes a spliceosome protein, have been identified in myeloid malignancies.27 Although results showed significant association of these genes with ABO antigen weakness, mutations detected in these genes were confined to specific hot spot, and there was no difference in mutations detected from patients with or without ABO antigen weakness. Further study is required to validate the association of these genes with ABO antigen weakness as well as to investigate the mechanism of reduced antigen expression of those gene mutations. Similar to CEBPA, there might be distinct phenotype or clinical characteristics among patients with mutated NRAS, PTPN11, and U2AF1 according to the presence of ABO antigen weakness.
There are limitations of this study. ABO genotyping was done in a limited number of patients, thus the reduced antigen expression in some patients may be due to ABO subgroups. Although the association of some genes with the ABO antigen weakness was confirmed, the mechanism and role of each gene could not be investigated. In addition, the allelic status of CEBPA mutations detected from patients could not be tested due to lack of samples. The retrospective design of our study, the limited cases of antigen weakness associated with specific mutations observed, and the lack of functional validation are additional limitations. To address these limitations, further studies are warranted.
Unexpected reactions such as weak ABO antigen in ABO typing should be dealt with caution due to the risk of transfusing incompatible blood components, which may compromise the safety of the patients. Understanding clinical situations that can cause ABO antigen weakness would be helpful for the transfusion practice in field. Furthermore, by elucidating the factors associated with the ABO antigens weakness, it will aid in understanding the mechanisms related to ABO antigen expression. In this study, we described the factors associated with ABO antigen weakness. Not only did we confirm the association with hematologic malignancies, we also revealed genes related to the phenomenon, especially CEBPAdm.
Acknowledgments
The authors thank Biostatistics Collaboration Unit of Yonsei College of Medicine for advice on statistical analysis. The authors also appreciate the Medical Illustration & Design team, a member of Medical Research Support Services of Yonsei University College of Medicine, for their excellent support with the visual abstract.
Authorship
Contribution: S.J.C., S.S.K., and S.K. conceptualized the study and developed methodology; S.J.C., H.K.K., E.J.S., S.S.K., and S.S. curated data and performed analysis; S.S.K., S.-T.L., and S.K. administered the project and supervised the study; S.J.C., H.K.K., and S.S.K. drafted the manuscript; and S.J.C., H.K.K., E.J.S, S.S.K., S.S., S.-T.L., and S.K. reviewed and edited the manuscript and approved its final version.
Conflict-of-interest disclosure: S.-T.L. is currently employed by Dxome. The remaining authors declare no competing financial interests.
Correspondence: Soon Sung Kwon, Department of Laboratory Medicine, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seoul 03722, Korea; email: kwonss@yuhs.ac.
References
Author notes
S.J.C. and H.K.K. contributed equally to this study.
Data are available upon reasonable request from the corresponding author, Soon Sung Kwon (kwonss@yuhs.ac).
The full-text version of this article contains a data supplement.