Key Points
Structure of mouse TPO bound to MPL illustrates ligand induces receptor dimerization and shows how patient mutations affect signaling.
MD modeling of JAK2 bound to MPL/TPO complex vs apo MPL reveals how MPL dimerization or patient mutations can induce JAK2 phosphorylation.
Visual Abstract
Myeloproliferative leukemia protein (MPL), also known as thrombopoietin (TPO) receptor, is a class I cytokine receptor that is expressed on hematopoietic progenitors, promoting growth and differentiation toward the megakaryocyte lineage and is critical for normal platelet production. Mutations in MPL, TPO, or Janus kinase 2 (JAK2) have been implicated in multiple diseases from congenital thrombocytopenias to myeloproliferative neoplasms. The ligand for MPL, TPO, stimulates platelet production by inducing MPL dimerization and results in an active conformation that allows downstream JAK2/STAT5 signaling. Despite the biological importance of this pathway, the molecular signaling mechanism remained unclear. Here, we present a 3.39-Å cryo-electron microscopy structure of the ectodomain of mouse MPL bound to TPO. The structure revealed both low and high affinity sites between MPL and TPO that contain several pathologic mutations. To better understand TPO-driven MPL signaling, we expanded upon this structure by molecular dynamic (MD) simulations to model the full-length human MPL/TPO complex, and showed that MPL D4-D4 domain interactions are functionally relevant in activity assays. To build on our understanding of downstream activation, we added JAK2 to the MPL/TPO complex by MD simulations. This ternary complex illustrates JAK2 dimerization through the pseudokinase domain, illustrates residues important for MPL interactions, and reveals the constitutive activation mechanism of patient mutant V617F. The model also suggests the mechanism of JAK2 tyrosine kinase domain transphosphorylation. Overall, our studies illuminate TPO/MPL/JAK2 signaling mechanisms and provide additional insight into the nature of receptor signaling, which will further benefit human health.
Introduction
Myeloproliferative leukemia protein (MPL) is a class I cytokine receptor that is expressed on hematopoietic stem cells and in the megakaryocyte lineage, and signals through Janus kinase (JAK) proteins in JAK/STAT pathways. MPL is a primary growth factor receptor on hematopoietic stem cells and contributes to the growth and differentiation of hematopoietic cells into megakaryocytes. Although MPL−/− does not lead to complete bone marrow failure in mice,1 lack of appropriate MPL signaling by either thrombopoietin (TPO) or MPL knockout leads to significant defects in proliferation of progenitors in the bone marrow.2
TPO is the ligand for MPL and is constitutively produced in the liver.3-6 TPO levels are regulated by binding to MPL on platelets that act as a sink for circulating TPO. When platelets are low, there is less MPL to bind and circulating TPO is high, inducing megakaryocytes to produce more platelets. TPO activates MPL by binding via a high-affinity interface (with a dissociation constant [KD] of 5-10 nM for human and 100-160 nM for mouse), which then recruits a second MPL molecule to its low-affinity interface (KD of 0.9 μM for human, and >10 μM for mouse) resulting in a productive signaling dimer (overall KD of 0.3 nM for human, and 1 nM for mouse).7-9 Diseases result from both loss-of-function and gain-of-function mutations in the MPL/TPO pathway. Patients with congenital amegakaryocytic thrombocytopenia (CAMT) are thrombocytopenic in spite of high TPO levels because of poor TPO/MPL binding, whereas activating mutants are associated with essential thrombocytosis that is characterized by excessive production of platelets (Table 1).10
TPO has been extensively studied for its role in hematopoietic stem cell maintenance, megakaryocyte differentiation, and platelet production but the molecular mechanism for this process is unclear. To elucidate the molecular basis for this process, we resolved the cryo-electron microscopy (cryo-EM) structure of the ectodomain of mouse MPL (ECD) complexed with the mouse TPO cytokine core domain (hereafter called mMPL/mTPO), which is conserved with human (hMPL/hTPO) proteins (79.5% and 83.2% similarity, respectively, supplemental Figures 1 and 2). mMPL binds mTPO in a 2:1 stoichiometry with high and low affinity binding sites. Our structure reveals that many patient mutations are in MPL/TPO interfaces and identifies other critical residues. While our work was in preparation for publication, other groups reported structures of hMPL/hTPO and mMPL/mTPO complexes8,9 and our work agrees well with these structures. We additionally leveraged our cryo-EM structure to model full-length (FL) hMPL/hTPO by molecular dynamic (MD) simulations that suggest MPL D4–D4 interactions are important for TPO-dependent activation and provide molecular insight into the mechanism of the constitutive active patient MPL mutant S505N. We further recapitulated the pathway by modeling the hMPL/hTPO/JAK2 structure, which illustrates interactions between the MPL intracellular domain and JAK2, JAK2-JAK2 dimerization, and JAK2 transphosphorylation. Overall, our work provides molecular insight into the mechanism of normal platelet production and mutations that result in blood diseases.
Methods
For more detailed methods, please see the supplemental Data.
Protein expression and purification
Proteins were expressed from HEK293 cells. A construct containing a fusion of mMPL ECD with a tobacco etch virus (TEV) cleavage site, a fragment crystallizable (Fc) tag, and an octahistidine (8His) tag (mMPL(26-482)-TEV-Fc-8His) was coexpressed with a construct containing mouse TPO cytokine core domain with a TEV cleavage site and 8His tag (mTPO (1-184)-TEV-8His). The protein complex was purified by nickel affinity followed by TEV protease cleavage, ion exchange, and size-exclusion chromatography. Human hTPO cytokine core domain (residues 22-184) wild-type (WT) and mutants contained C-terminal DYKDDDDK (FLAG) tags and were purified with anti-FLAG resin and size-exclusion chromatography.
Cryo-EM
mMPL/mTPO at 1 mg/mL was vitrified with a Vitrobot Mark IV on Quantifoil 300 mesh R 1.2/1.3 holey carbon film grids. Cryo-EM videos were collected on a Titan Krios transmission electron microscope with a K3 detector. Data were processed with cryoSPARC 4.1.111 with the aid of Patch CTF. AlphaFold212 predicted mTPO (AF-P40226) and mMPL ECD (AF-Q08351) structures were used for initial fitting followed by manual adjustment and building in COOT13 and refinement with Phenix software14 (supplemental Table 1).
STAT5 reporter assay and MPL cell surface expression
Lentivirus transduction was used to generate the HEK293 STAT5-NanoLuc/STAT5b stable cell line. To monitor STAT5 activation, cells were transiently transfected with WT or mutant hMPL constructs. Transfected cells were treated with recombinant hTPO for 18 hours. Nano-Glo Luciferase and CellTiter-Glo luminescence reagents (Promega) were added to determine nanoluciferase activity and the number of viable cells, respectively. Flow cytometry was used to measure MPL surface expression. At 48 hours after hMPL construct transfection, cells were collected and stained with anti-CD110 allophycocyanin antibody (BioLegend). Flow cytometry data were acquired and FlowJo (BD Life Sciences) was used for data analysis.
MD simulations
AlphaFold2-based structure prediction15, computational homology modeling16, and molecular docking were used to develop the FL hMPL/hTPO and the FL hMPL/hTPO/JAK2 complexes. Modeled complexes underwent further refinement and analysis through extensive MD simulations, for 20 μs. For these MD simulations, each model construct was embedded in a membrane bilayer, immersed in water and ions, containing an excess of 0.15 M NaCl, which led to simulation boxes containing as many as 800 000 particles for full atomistic MD simulations. All MD simulations (supplemental Table 2) were conducted using the CHARMM36m force field,17 and the computations were executed with GROMACS-2022.18
Results
The cryo-EM structure of mMPL/mTPO reveals that patient mutations occur at high and low affinity interfaces, and the near the WGPWS motif
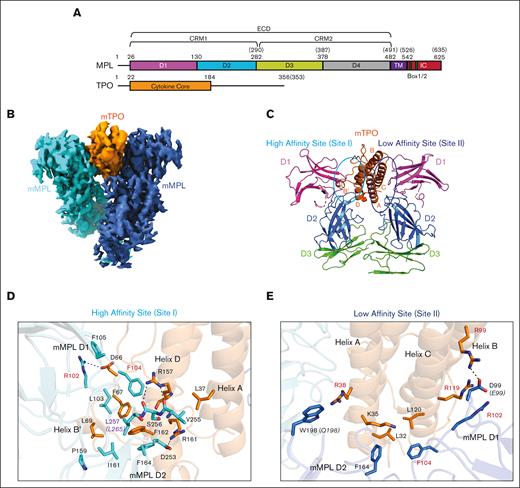
The MPL ECD is composed of 2 cytokine receptor modules (CRMs): CRM1, formed by domains D1 and D2, and CRM2, formed by domains D3 and D419,20 (Figure 1A, top). TPO is a secreted cytokine with a structured core domain that binds MPL ECD followed by a C-terminal, highly glycosylated extension (Figure 1A, bottom).21-24 We coexpressed and copurified mMPL ECD with mTPO cytokine core domain and reconstructed the complex structure to 3.39-Å resolution by cryo-EM (Figure 1B; supplemental Figures 3A-C and 4A-D; supplemental Table 1). The cryo-EM map showed 1 copy of mTPO sandwiched between 2 copies of mMPL ECD (Figure 1B). mTPO forms a 4-helical bundle similar to the crystal structure of hTPO bound to a neutralizing antibody7 (Figure 1C; supplemental Figure 5A). Two copies of mMPL domains D1 and D2 were fit into generally well-resolved map density and D3 domains were fit roughly into more diffuse density (supplemental Figure 5B-G). We observed additional partial density for a loop (residues 200-228) in the mMPL D2 domain near the low affinity site but did not fit this because of ambiguity of chain direction (supplemental Figure 6A). Density suggested that the 2 D4 domains approach but was not well resolved, as observed previously9 so D4 domains were not fit (supplemental Figure 6B). Our mMPL/mTPO complex was prepared in a manner that did not retain an engineered dimerization domain on mMPL or use crosslinking, in contrast to other reported structures,8,9 which may have resulted in more flexibility for our complex and lower local resolution in regions outside of the mMPL/mTPO interface. Despite this, the overall architecture of our structure is in good agreement with previously published mMPL/mTPO and hMPL/hTPO cryo-EM structures8,9 (supplemental Figure 6C and D, respectively). Mass spectrometry analysis revealed the expected N-glycosylation on mMPL residues N117, N178, and N349 (supplemental Figure 7), but glycans were not fit because of weak map density in these areas.
The D1 and D2 domains of both copies of mMPL ECD engage mTPO at high and low affinity sites, also known as sites I and II, respectively (Figure 1C). mMPL residues R102, L103, F104, and F164 make key interactions in the high affinity site (Figure 1D). R102 makes a salt bridge to mTPO D66 and stacks with mMPL F105. mMPL L103 hydrophobically interacts with mTPO L69, and mMPL F104 stacks between mTPO F67 and R157. mMPL F164 stacks with mTPO F162, hydrophobically interacts with mMPL I161, and stacks with mTPO F67. Additionally, a hydrophobic interaction between mTPO L37 and mMPL V255, and a hydrogen bond between mTPO R157 and mMPL S256 occur (Figure 1D). Importantly, R102 and F104 (Figure 1D, red text) are both mutated in CAMT (Table 1), indicating a clear connection between residues important for TPO interaction and loss of signaling function. Conserved mMPL residue L257 (Figure 1D, purple text, L265 in human) near a hydrophobic interface is required for TPO binding in both mouse25 and human,26 and the L265F patient mutation results in increased signaling and high platelet counts.27 Computational modeling of the L257F mutation indicates closer distances to conserved mTPO residues F162 and F167, and mMPL residues L103 and I161 (supplemental Figure 8), suggesting a stronger interaction and a plausible mechanism for increased signaling.
In the low affinity site, mTPO R99, R119, and R38, and mMPL R102 and F104 that are mutated in CAMT (Table 1; Figure 1E, red text) are involved in interfaces. mTPO R99 makes a salt bridge to mMPL D99, mTPO R119 stacks with mMPL R102, and mTPO L32 and L120 hydrophobically interact with mMPL F104 (Figure 1E). mTPO R38 interacts with nonconserved mMPL W198, and mMPL F164 hydrophobically interacts with mTPO L32 and K35 (Figure 1E). Notably, mMPL residues R102, F104, and F164 all also make key interactions in the high affinity site (Figure 1D).
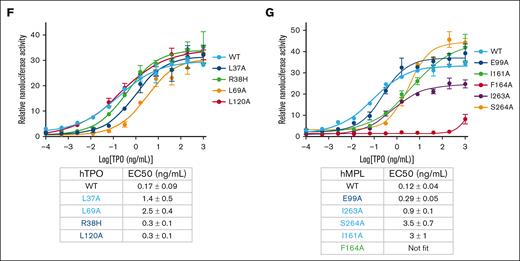
Our structure revealed that several residues that were previously shown to be critical for receptor signaling are in the MPL/TPO interfaces and agrees with results from previous biochemical studies.28-31 A few key residues, such as MPL F164, to the best of our knowledge, had not been previously investigated by mutagenesis. To investigate a system more related to disease states, we used mutagenesis and a reporter assay to test the importance of these residues in the function of hMPL/hTPO interactions. We used a HEK293 cell line stably expressing both STAT5b and the STAT5 response element upstream of a NanoLuc reporter element. Cells were transfected with FL hMPL plasmid and recombinant hTPO core domain protein was added to induce response. All hTPO mutants resulted in a loss of signaling efficiency, but the effect was greater for mutants in the high affinity site (∼8-15-fold in 50% effective concentration) vs the low affinity site (<2-fold in 50% effective concentration; (Figure 1F) compared with WT hTPO. hMPL mutations were tested in the same format with the addition of flow cytometry analysis because many hMPL mutants feature reduced cell surface expression.25,32,33 All hMPL mutants had comparable or higher expression on the surface to WT except I263A (supplemental Figure 9), suggesting that results from this mutation may not be solely because of reduced hTPO binding. Mutation of residues that were not strictly conserved between mouse and human, E99 (low affinity site, D99 in mouse) and I263 (high affinity site, V255 in mouse), resulted in ∼2.4- to 7.5-fold reduction in activation (Figure 1G). In contrast, mutation of conserved residues I161 and S264 (S256 in mouse), in the high affinity site, resulted in ∼25- to 29-fold reduction in activation, and mutation of F164 that binds to both TPO sites abolished activation (Figure 1G). Together, mutagenesis and functional assays validated our structure and highlighted residues that are critical for MPL–TPO interaction in mouse and human.
mMPL contains the WGPWS sequence (residues 261-265) characteristic of class 1 cytokine receptors19,20 in the D2 domain that forms a π-stacking arginine-tryptophan ladder composed of W261, R249, W264, R170, and W245 that is in proximity to the N-terminus of mMPL (supplemental Figure 10A-B). Mutation of human residue R257 corresponding to mMPL R249 results in loss of signaling in CAMT.34-36 R249 stacks with W261 that is downstream of residues 253 to 258 that form part of the high affinity interface, suggesting long-range destabilization may result from mutation (supplemental Figure 10A). WXXWS is a mannosylation consensus sequence but only 10% of our mMPL protein was mannosylated at residue W261 (supplemental Figure 10C-D), yet additional mannosylation in this region was reported in hMPL.8,37 Notably, the first residue in the arginine tryptophan ladder, W245, is next to Y244 that, in humans, results in increased megakaryocyte growth upon TPO stimulation when mutated (Y252H),38 suggesting other possible connections between disease states and the integrity of this motif (Table 1).
A D4-D4 interaction is revealed by computational modeling of FL hMPL/hTPO complex
The MPL transmembrane (TM) and intracellular (IC) regions (Figure 1A) are known to be important for its dimerization and activation, but our cryo-EM mMPL/mTPO structure only visualized the mMPL ECD without the D4 domains fit. Although we note that D4 domains are included in other hMPL/hTPO and mMPL/mTPO structures, issues with precise fitting were also indicated in the latter.8,9 Given 2 CRMs of MPL rather than 1 in other family members, we wanted to further understand their role in TPO-induced MPL activation and used MD simulations to characterize the FL hMPL/hTPO complex. We performed rigorous computational modeling steps (supplemental Figures 11-13), used homology modeling using our cryo-EM structure as the initial template, and further used the AlphaFold predicted FL hMPL model (AF-P40238-F1-model_v4) to incorporate the remaining domains (Figure 2A).
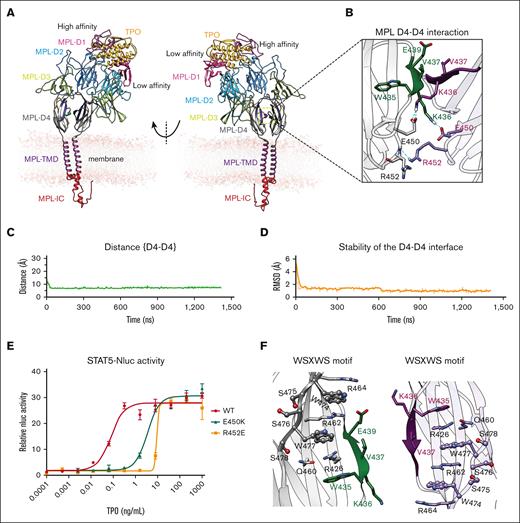
hMPL D4-D4 interaction stabilizes the hMPL dimer upon hTPO binding. (A) The optimized computational modeling structure of the FL hMPL dimer bound to hTPO, embedded in a lipid bilayer membrane, as derived from a ∼1.5 μs MD simulation. (B) Detailed view of the intermolecular interactions formed at the identified D4-D4 interface in the activated MPL dimer. Hydrogen bonds are represented by cyan dashed lines. The stability analysis of interaction at the D4-D4 interface measured as: (C) the interdomain distance between D4-D4, considering the center of mass of the Cα atoms within residues 435-440. These residues delineate the antiparallel β-sheet, formed at the interface between the 2 hMPL D4 domains; and (D) the RMSD analysis, within residues 435-440 for both hMPL chains that define the antiparallel β-sheet at the interface. The RMSD was computed for all heavy atoms (nonhydrogen atoms), including side chains. For this analysis, each simulation snapshot was compared against the final configuration derived from the ∼1.5-μs MD simulation. (E) Reporter assay using transfected human FL hMPL WT and D4-D4 mutant with recombinant WT hTPO core domain. A representative result is shown with error bars representing the standard deviation of technical replicates. The table shows the mean ± standard deviation from 3 independent experiments. (F) The WSXWS motif in the D4 domain (gray and light violet) contributes to preserving the structural stability of the established D4-D4 interface.
hMPL D4-D4 interaction stabilizes the hMPL dimer upon hTPO binding. (A) The optimized computational modeling structure of the FL hMPL dimer bound to hTPO, embedded in a lipid bilayer membrane, as derived from a ∼1.5 μs MD simulation. (B) Detailed view of the intermolecular interactions formed at the identified D4-D4 interface in the activated MPL dimer. Hydrogen bonds are represented by cyan dashed lines. The stability analysis of interaction at the D4-D4 interface measured as: (C) the interdomain distance between D4-D4, considering the center of mass of the Cα atoms within residues 435-440. These residues delineate the antiparallel β-sheet, formed at the interface between the 2 hMPL D4 domains; and (D) the RMSD analysis, within residues 435-440 for both hMPL chains that define the antiparallel β-sheet at the interface. The RMSD was computed for all heavy atoms (nonhydrogen atoms), including side chains. For this analysis, each simulation snapshot was compared against the final configuration derived from the ∼1.5-μs MD simulation. (E) Reporter assay using transfected human FL hMPL WT and D4-D4 mutant with recombinant WT hTPO core domain. A representative result is shown with error bars representing the standard deviation of technical replicates. The table shows the mean ± standard deviation from 3 independent experiments. (F) The WSXWS motif in the D4 domain (gray and light violet) contributes to preserving the structural stability of the established D4-D4 interface.
Interestingly, in our MD FL hMPL/hTPO complex a D4-D4 interface forms composed of an antiparallel β-sheet structure within residues 434 to 440 (Figure 2B-C) and exhibits a root mean squared deviation (RMSD) of 1.8 Å with minimal change throughout simulation (Figure 2D). Prominently, E450 forms a salt bridge with K436, whereas R452 hydrogen bonds with R452 on the opposite chain, collectively stabilizing the interface (Figure 2B). E450K and R452E mutations resulted in 33-fold and 92-fold lower signaling, respectively, than WT MPL in the STAT5 reporter assay (Figure 2E), with no defect in cell surface expression (supplemental Figure 9), highlighting the importance of this interaction in productive MPL signaling. The model also revealed the WSSWS motif (residues 474-478, WSASW, and corresponding to residues 465-468 in mMPL) significantly contributes to maintaining the structural integrity of the D4–D4 interface (Figure 2F). This motif is mannosylated in hMPL8,37 (W473) and mMPL (W465), as previously reported.9
Although the hMPL dimer remains closed and tightly bound to hTPO in MD simulation replicates, there is variability resulting in a range of conformational states in solution, from upright to slightly bent, reflecting its dynamic nature and the flexibility of the CRM regions (Figure 2A; supplemental Figure 14).
The dimerization and activation of hMPL constitute a hTPO ligand-induced process
To further understand the importance of TPO binding to MPL receptor activation, we removed hTPO from our FL hMPL/hTPO model and conducted 3 long-timescale MD simulations (∼8 μs) (Figure 3A; supplemental Figure 15A), resulting in a substantial expansion of the ECD-D1 domains (Figure 3A). The interdomain D1-D1 separation enlarges dramatically, shifting from an initial 60 Å in the activated complex (Figure 3B, top) to 120 Å, thereby adopting an open conformation. Notably, 1 of our replicate simulations indicates that hMPL can also form an overly compact dimer, with D1-D1 distance shrinking to 25 Å (supplemental Figure 15A), a state that would be unfavorable for hTPO binding.
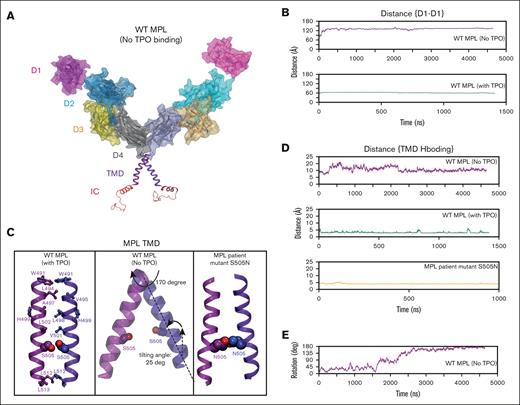
The productive dimerization of hMPL, induced by hTPO binding. (A) The optimized configurations of hMPL dimer without hTPO presence. (B) Distance analysis of the D1-D1 domain indicating that the hMPL ECD adopts open configurations when hTPO is absent. Here, the distance between the center of mass of Cα atoms within residues (26-130) was measured across both hMPL D1 domains. (C) The configurations of the TMD-TMD interface under various conditions: hTPO-bound WT hMPL dimer, unbound (apo) WT hMPL dimer, and unbound (apo) S505N mutant hMPL dimer. The formation of an S505 hydrogen bond with its counterpart on the adjacent hMPL leads to the activation of the dimer. (D) Analysis of the hydrogen bonding between TMD-TMD at position 505 in the TPO-bound WT hMPL dimer, the unbound apo WT hMPL, and the unbound hMPL S505N dimers. The distance between oxygen gamma (OG) atoms on S505-S505 interaction and between carbon gamma (CG) atoms on N505-N505 interaction was considered for this analysis. (E) Without hTPO binding, the TMD shifts into nonproductive configurations due to the disruption of the hydrogen bonding between S505-S505, leading to a 170-degree rotation of the TM around its axis.
The productive dimerization of hMPL, induced by hTPO binding. (A) The optimized configurations of hMPL dimer without hTPO presence. (B) Distance analysis of the D1-D1 domain indicating that the hMPL ECD adopts open configurations when hTPO is absent. Here, the distance between the center of mass of Cα atoms within residues (26-130) was measured across both hMPL D1 domains. (C) The configurations of the TMD-TMD interface under various conditions: hTPO-bound WT hMPL dimer, unbound (apo) WT hMPL dimer, and unbound (apo) S505N mutant hMPL dimer. The formation of an S505 hydrogen bond with its counterpart on the adjacent hMPL leads to the activation of the dimer. (D) Analysis of the hydrogen bonding between TMD-TMD at position 505 in the TPO-bound WT hMPL dimer, the unbound apo WT hMPL, and the unbound hMPL S505N dimers. The distance between oxygen gamma (OG) atoms on S505-S505 interaction and between carbon gamma (CG) atoms on N505-N505 interaction was considered for this analysis. (E) Without hTPO binding, the TMD shifts into nonproductive configurations due to the disruption of the hydrogen bonding between S505-S505, leading to a 170-degree rotation of the TM around its axis.
The elimination of hTPO reverberates through the D4-D4 and TM-TM interfaces, disrupting interactions that were established in the activated complex (supplemental Figure 15B; Figure 3C). These calculations show that hMPL cannot stably homodimerize to be active without hTPO association. In contrast, the activated hMPL dimer after hTPO binding has a strong TM-TM interface mediated by tight interdomain interactions including the D4-D4 interaction (Figures 2A and 3B) and notably a hydrogen bond between S505 residues (Figure 3C, left). The S505 hydrogen bonding orients the helices into a left-handed coiled-coil arrangement, in good agreement with the proposed model of activated TM dimer obtained from a nuclear magnetic resonance (NMR) study.39,40 In the absence of hTPO, hydrogen bonding between the S505 residues breaks (Figure 3C, middle), changing the distance from ∼3 to 4 Å to 10 to 20 Å (Figure 3D, top) leading to dissociation of the TMs and a 170° rotation (Figure 3E). Rotation of TM helices around their axis determines 2 distinct conformations: “histidine-in,” which is activated, and “histidine-out,” which is inactive, also identified in previous MD simulations.41,42 Interestingly, we also noticed that the rotation around the axis is accompanied by a 25° tilting of the TM helices relative to the bilayer normal. Measurements using polarized infrared spectroscopy43 show that the hMPL dimerization is disrupted by increasing the tilt angle of the helix relative to the bilayer normal. Pronounced tilting of TM helices is also evident in a published computational modeling structure44 in which the ECD domains and TPO were absent. Overall, our model shows that upon the dissociation of hTPO the structural integrity of the hMPL dimer is lost and the complex is likely to break down.
hMPL residue S505N is mutated in patients with familial essential thrombocythemia (ET), a condition believed to result from autonomous homodimerization,45 and was shown to result in constitutive activity.39 Our MD simulations for MPL S505N indicate that unlike the WT apo hMPL, the TMD conformation of the S505N mutant remains in the activated state (Figure 3C, right), largely because of the formation of an interdomain hydrogen bond between the N505 residues (Figure 3C right and Figure 3D bottom; supplemental Figure 16). We reexamined this calculation in the presence of activated JAK2. Our analysis indicates that although the N505 hydrogen bonding in hMPL remains intact, it also stabilizes JAK2 dimerization through the pseudokinase (PK)-PK interaction (supplemental Figure 16B), which is crucial for subsequent signaling events.44
MPL dimerization supports JAK2 dimerization and trans activation
We modeled a JAK2 dimer bound to hMPL (Figure 4A) to better understand the hTPO-mediated mechanism of downstream hMPL signaling. The efficient recruitment and dimerization of JAK2 are coordinated through tight interactions between the IC-Box1/Box2 domains of hMPL, and the band 4.1-ezrin-radixin-moesin (FERM) and SH2 domains of JAK2 (Figure 4B-C; supplemental Figure 17A,D-E). Our model indicates that hMPL Box1 domain residues W529, P530, S531, L532, P533, and D534 engage in hydrophobic interactions with JAK2 FERM domain (Figure 4B; supplemental Figure 17D) in agreement with previous studies indicating the importance of the interface between the JAK2 FERM domain and hMPL Box1 region.46 Our MD simulations also reveal that the hMPL Box2 region residues L567-L571 interact with the JAK2 SH2 domain (Figure 4C; supplemental Figure 17E).
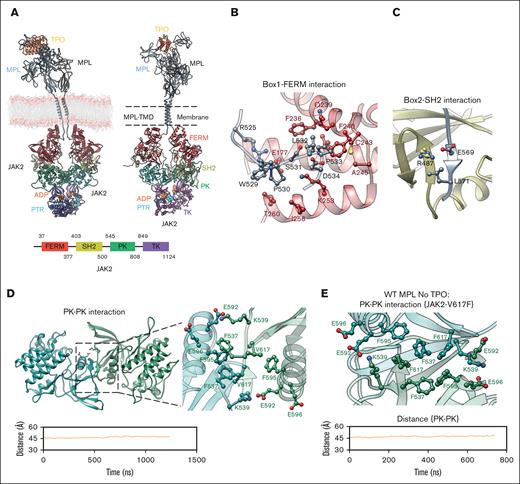
JAK2 dimerization mechanism. (A) The computational model of the FL hMPL-hTPO-JAK2 complex embedded in a lipid bilayer membrane. Left: the modeling construct before MD simulations; right: the system after a 1.3-μs MD simulation; bottom: JAK2 domain scheme with each domain color coded. (B) Interaction between the JAK2 FERM domain (red) and hMPL Box1 domain (gray). (C) Interaction between the JAK2 SH2 domain (yellow) and hMPL Box2 domain (gray). (D) JAK2 dimerization via trans PK-PK interaction. Top: stability analysis of the PK-PK (green) interface in the presence of hTPO, determined by measuring the distance between the center of mass of Cα atoms within residues (530-540 and 545-808) across both JAK2 PK domains. The bottom panel provides a detailed view of the intermolecular interactions between 2 PK domains in the FL WT hMPL-hTPO-JAK2 system. (E) JAK2 V617F constitutive activity mechanism. Top: detailed intermolecular view of the PK-PK interface with V617F mutation within the apo hMPL-JAK2 structure. Bottom: stability analysis of apo hMPL PK-PK interface. The introduction of F617 leads to the formation of π-π stacking interactions. All key residues are highlighted as sticks.
JAK2 dimerization mechanism. (A) The computational model of the FL hMPL-hTPO-JAK2 complex embedded in a lipid bilayer membrane. Left: the modeling construct before MD simulations; right: the system after a 1.3-μs MD simulation; bottom: JAK2 domain scheme with each domain color coded. (B) Interaction between the JAK2 FERM domain (red) and hMPL Box1 domain (gray). (C) Interaction between the JAK2 SH2 domain (yellow) and hMPL Box2 domain (gray). (D) JAK2 dimerization via trans PK-PK interaction. Top: stability analysis of the PK-PK (green) interface in the presence of hTPO, determined by measuring the distance between the center of mass of Cα atoms within residues (530-540 and 545-808) across both JAK2 PK domains. The bottom panel provides a detailed view of the intermolecular interactions between 2 PK domains in the FL WT hMPL-hTPO-JAK2 system. (E) JAK2 V617F constitutive activity mechanism. Top: detailed intermolecular view of the PK-PK interface with V617F mutation within the apo hMPL-JAK2 structure. Bottom: stability analysis of apo hMPL PK-PK interface. The introduction of F617 leads to the formation of π-π stacking interactions. All key residues are highlighted as sticks.
Furthermore, hMPL mutations such as E569A, I570A, and L571A significantly decrease JAK2 binding.44 This interaction induces the hMPL Box2 region to form an antiparallel β-sheet. The similar structural configuration was revealed by the cryo-EM structure of interferon γ receptor 1 (IFNγR1)/mJAK1.47,48 Notably, within this hMPL/JAK2 interaction interface, a salt bridge between hMPL E569 and JAK2 R487 emerges as a key point of connection (Figure 4C). Thus, our hMPL/hTPO/JAK2 model provides additional structural insights into how hMPL engages JAK2 to transduce signaling.
In addition, our model highlights the interactions between hMPL and JAK2 within the juxtamembrane domain (supplemental Figure 18), characterized by hydrophobic contacts between hMPL W515 and residues I223, L224, and K227 on the FERM domain. Notably, a direct interaction between W515 and L224 was observed, which resides on the membrane surface and plays a pivotal role in anchoring JAK2 to the membrane.44 W515 undergoes a significant conformational change during activation. In the inactive hMPL structure, W515 is involved in an internal cation-π interaction with R514. Upon hTPO binding, this interaction is disrupted, enabling W515 to rotate toward JAK2. This rotation leads to the unraveling of the α-helix in the W515-Q516-F517-P518 region. Solid-state NMR49 associates this structural transformation with hMPL activation and the disruption of the internal W515-R514 cation-π interaction, facilitating W515 interaction with the FERM domain. An engineered mutation, L224E, designed to disrupt the FERM interaction with the membrane, decreased JAK2 dimerization and subsequent signaling.44 Importantly, W515L/K mutations are linked to primary (idiopathic) myelofibrosis and ET50-52 so the L224-W515 connection could be of note for future studies.
The dimerization of JAK2 is primarily mediated via PK–PK interface interactions in a trans configuration, which remains stable at 45 Å throughout MD simulations. The tight PK-PK interface is formed through symmetric hydrophobic interactions involving V617, F537, and F595, coupled with ionic interactions extending from K539 to E592/E596 (Figure 4D). These interactions induce the formation of an antiparallel β-sheet structure within M535-H538. A similar interface was seen in the cryo-EM structure of mJAK1 and miniIFNλR1.47,48 The significance of the PK-PK interface is further emphasized by the fact that truncating the entire PK disrupts MPL/JAK2 dimerization and signaling.44 The polycythemia vera–associated V617F mutation at the PK-PK interface induces constitutive activity in JAK2.53-56 To better understand this mechanism, we conducted a 600-ns MD simulation on the hMPL/JAK2 excluding hTPO that revealed significant differences between the hMPL-hTPO-JAK2 complex and hMPL-JAK2(V617F). Notably, in the latter structure (supplemental Figure 19), the hydrogen bond at S505 in the TMD tends to break, aligning with a recent report.57 This indicates that the TM in the hMPL-JAK2(V617F) structure undergoes a substantial shift, adopting a configuration that is rotated ∼110° relative to the TM of hMPL-hTPO-JAK2. Although the limited timescale in our study prevented us from directly observing this rotational shift, we earlier showed (Figure 3C,E) that upon breaking the hydrogen bond between S505-S505, the TM of the hMPL dimer rotates significantly (by ∼170°) to adopt a structure markedly different from the fully activated state. This suggests that a similar rotation is also expected to occur in hMPL-JAK2(V617F). This rotation, along with the wide opening in the ECD (supplemental Figure 19), results in a structure that is markedly different from that observed in the fully activated state of hMPL-hTPO-JAK2. Despite significant structural rearrangement, V617F in JAK2 maintains the stability of the PK-PK trans interface (Figure 4E). V617F engages in π-π stacking with F595 and F537, leading to a dense interaction network that does not require hTPO binding to maintain dimerization. Analogous to the V617F mutation in JAK2, the V657F mutation in mouse JAK1 has been elucidated by cryo-EM.48 V657F promotes constitutive dimerization of mJAK1 by facilitating a tight interface between the PK domains, primarily driven by strong π-π stacking interactions. Such constitutive activity is associated with acute leukemia observed in patients with hJAK1 V658F.58 We also observe ionic interactions between K539 and E592/E596 in both mutant and WT complex.59,60 Supporting this, mutations at the PK-PK interface, including F595A, E596R, and K539E all resulted in the suppression of JAK2 V617F hyperactivity.59-61
Productive JAK2 dimerization triggers transphosphorylation at the TK domain
JAK2 is phosphorylated at Y1007 and Y100862 in the tyrosine kinase (TK) activation loop, so we modeled these as the phosphorylated forms. Although the AlphaFold2 models47,63 present an interface between TK-TK domains, the position of the TK activation loop does not offer a comprehensive understanding of activation and phosphorylation mechanisms.
In our model, 1 TK domain forms a strong interface with its neighboring TK domain and maintains stability at a distance of 44 Å, long enough to facilitate transphosphorylation on Y1007 and Y1008, hereafter called PTR1007 and PTR1008 (Figure 5). We see a stable interaction that allows the activation loop to reach the nucleotide binding pocket (Figure 5) of the adjacent monomer, allowing this phosphorylation event. Interestingly, even starting with the adenosine diphosphate (ADP)-bound JAK2 in our model, which includes PTR1007 and PTR1008, the activation loop displays an inclination to approach ADP, with K1009 primary mediating this binding. A pronounced interaction between PTR1007, PTR1008, and K1009 with ADP is evident (Figure 5). This scenario could be interpreted as the commencement of the phosphorylation process. Our analysis suggests that a dominant electrostatic attraction between K1009 and ADP drives this movement, leading to notable interactions between the activation loop and ADP. Notably, the K1009A mutant was previously shown to result in reduced phosphotyrosine levels,64 in agreement with its prominent role in our model.
JAK2 activation mechanism.Trans phosphorylation of JAK2 TK (purple) domain. Stability analysis of the TK-TK interface, measured by the distance between the center of mass of Cα atoms within residues (849-1124) in both JAK2 TK domains. The right panel displays the activation loop of 1 TK domain penetrating the nucleotide-binding pocket of the adjacent TK domain, elucidating the mechanism of JAK2 activation. The solid line on the graph shows the exponential moving average over the course of a 1.3-μs MD simulation.
JAK2 activation mechanism.Trans phosphorylation of JAK2 TK (purple) domain. Stability analysis of the TK-TK interface, measured by the distance between the center of mass of Cα atoms within residues (849-1124) in both JAK2 TK domains. The right panel displays the activation loop of 1 TK domain penetrating the nucleotide-binding pocket of the adjacent TK domain, elucidating the mechanism of JAK2 activation. The solid line on the graph shows the exponential moving average over the course of a 1.3-μs MD simulation.
We propose that the inherent flexibility of the TK domain is vital for MPL-JAK2–mediated signaling. This adaptability, also discernible in the cryo-EM structures of mJAK1,47,48 suggests a dynamic range of possible conformations. The ML-based structure reported earlier,47 might depict just 1 of many relevant configurations (as in Figure 4A) leading to transphosphorylation. This dynamic nature of the TK domain is likely essential, enabling the targeting of remote tyrosine residues for phosphorylation, such as those on the MPL IC region, critical for STAT protein signaling.48,65 Our MD simulation suggests that the increased flexibility of the TK domain (supplemental Figure 20A) after transphosphorylation could potentially lead to the phosphorylation of accessible tyrosine residues, such as Y570 (supplemental Figure 20B) and Y637 (supplemental Figure 20C). These residues are associated with the autoinhibition65 and optimal JAK2 function,66 respectively.
Discussion
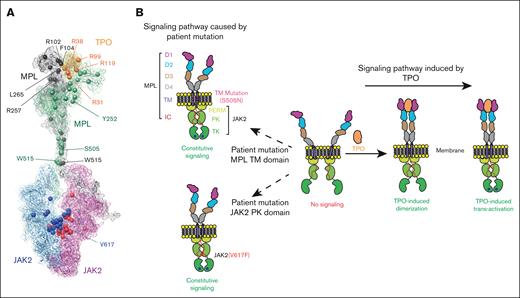
We report the cryo-EM structure of the mMPL/mTPO complex that reveals several patient mutations are found at interfaces (Figure 6A) and agrees with a prior reported structure9 whereas our MD-simulation structure agrees with the cryo-EM hMPL/hTPO structure8 (supplemental Figure 21). Our modeling of the FL hMPL/hTPO complex reveals D4-D4 interactions that were functionally validated and stabilize the dimer in the active conformation, and key residues in the TM domain. Like its close relative the erythropoietin receptor, (supplemental Figure 22), MPL is a homodimer with prebound JAK2 on each cytoplasmic region bound to Box1 and Box 2. The JAK2 proteins are likely largely in an inactive conformation that requires phosphorylation of an activation loop to stabilize the active conformation.48
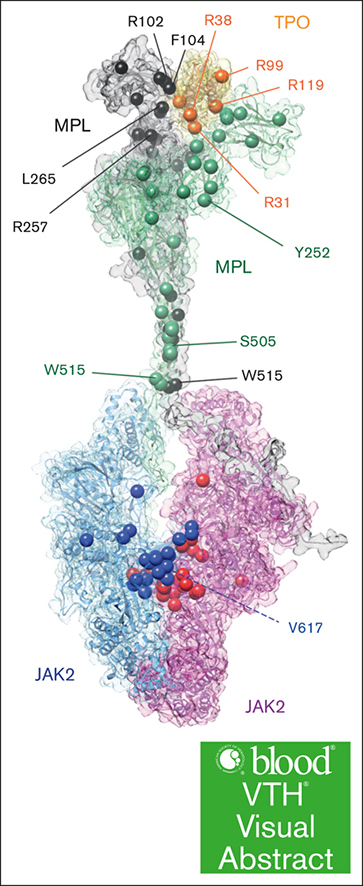
Model of hTPO-hMPL-JAK2 axis pathway. (A) Map of patient mutations in the hMPL/hTPO/JAK2 complex. The patient mutations are highlighted by spheres. (B) Schematic depiction of the hMPL dimerization and activation, as well as JAK2 association and activation mechanism facilitated by TPO binding.
Model of hTPO-hMPL-JAK2 axis pathway. (A) Map of patient mutations in the hMPL/hTPO/JAK2 complex. The patient mutations are highlighted by spheres. (B) Schematic depiction of the hMPL dimerization and activation, as well as JAK2 association and activation mechanism facilitated by TPO binding.
Our findings reveal a multifaceted picture of MPL activation (Figure 6B) and dimerization. When bound to TPO, the activated MPL dimer adopts a compact conformation in ECD (with the critical D4-D4 interface formation) and a closed TMD configuration (facilitated by S505-S505 interdomain hydrogen bonding) to trigger proper downstream signaling. In contrast, the apo MPL displays a vastly expanded ECD-D1 domain, a conformational change that propagates through to result in an open, nonproductive TMD. This observation aligns with a previous single-molecule fluorescence microscopy study, which reported very low levels of MPL dimerization in the absence of TPO ligand.44 Remarkably, the MPL S505N patient mutant diverges from this trend, as its TMD maintains a closed, constitutively activated configuration because of making a stronger hydrogen bond.
TPO binding induces the activation of the MPL dimer, which in turn efficiently leads to JAK2 dimerization. The dimerization of JAK2 is facilitated by a stable and tight interaction within the PK-PK domain, forming a trans configuration essential for JAK2 signaling. The V617F mutation contributes to a tight π-π stacking network, stabilizing the PK-PK interface, and leading to constant MPL/JAK2 activation in the absence of TPO. Our findings indicate the stable PK-PK interface provides sufficient time for the TK domain to align with its adjacent counterpart. This prolonged interaction allows the activation loop to access the nucleotide pocket in nearby JAK2 molecules, culminating in phosphorylation. Our findings enhance the understanding of TPO/MPL/JAK2 signaling, complement existing experimental data, and provide insights for future therapeutic research in hematopoietic growth factor signaling.
Acknowledgments
The authors thank Houxia Shi and Wenjun Gui and the team from Biortus Biosciences for the mMPL/mTPO complex preparation. They thank Yuan Qiao and Jack Yan from Biortus Biosciences for help on cryo-EM data collection and processing; Crystal Ghosh from Calico Life Sciences for project management; and Bob Cohen, David Stokoe, and Jonathan Powell from Calico Life Sciences for reviewing the manuscript and providing suggestions. They also thank Kailyn Kong, Cillian Variot, and John C. K. Wang from Calico Life Sciences for help with protein expression.
Authorship
Contribution: A.M. performed MD based simulations and analyzed the structure models; M.B. designed protein expression constructs and supervised protein production efforts, refined and finalized the mMPL/mTPO cryo-EM structure, and analyzed the structure to design mutations for functional assays; J.G., A.A.B., and D.H. generated constructs and performed STAT5 reporter assays; J.S. expressed and purified hTPO proteins; M.K.R. analyzed data; A.T.D., D.N., and N.J.B. performed glycan analysis by mass spectrometry; Q.H. and M.P. designed and supervised the experiments with additional supervision by Y.K. and D.E.; and A.M., M.B., Q.H., and M.P. wrote the manuscript with contributions and revisions from all authors.
Conflict-of-interest disclosure: A.M., M.B., M.K.R., J.S. D.E., Q.H., and M.P. are employees of Calico Life Sciences. LLC. J.G., A.A.B., D.H., A.T.D., D.N., N.J.B., and Y.K. are employees of AbbVie and may hold AbbVie stock.
Correspondence: Qi Hao, Calico Life Sciences LLC, 1130 Veterans Blvd, South San Francisco, CA 94080; email: qhao@calicolabs.com; and Marcia Paddock, Calico Life Sciences LLC, 1130 Veterans Blvd, South San Francisco, CA 94080; email: marcia@calicolabs.com.
References
Author notes
A.M. and M.B. contributed equally to this study.
Q.H. and M.P. are joint senior authors.
Coordinate and map files for the mouse MPL/mouse TPO structure were deposited in the Protein Data Bank and Electron Microscopy Data Bank under accession codes pdb 8vu5 and EMD-43526, respectively.
The full-text version of this article contains a data supplement.