Deep-sequencing technologies have identified a growing list of genes that are clinically significant in hematologic malignancies and therefore incorporated into heme-directed next-generation sequencing panels. However, with a few notable exceptions, most of these gene mutations are not directly targetable. Therefore, identification of new targetable lesions is a cause for excitement. Molecular genetic lesions involving the NTRK family of genes have been shown in recent studies to be involved in rare cases of hematologic malignancies. NTRK1, NTRK2, or NTRK3 genes encode the tropomyosin-related kinase (Trk) receptors TrkA, TrkB, or TrkC, respectively.1 Trk activation results in receptor dimerization and phosphorylation, followed by stimulation of downstream pathways including JAK/STAT, PI3K/AKT, and RAS/MAPK to promote proliferation, differentiation, and survival.1 NTRK fusions (with a variety of partners) are well-characterized oncogenic drivers of various solid tumors, found in nearly all cases of rare neoplasms such as congenital fibrosarcoma, secretory breast carcinoma, and mammary-analog secretory carcinoma of the salivary glands. Rarely, they are also found in much more common adult and pediatric tumors including lung, colon, melanoma, breast, pancreatic, and many others.2
Basket trials, which are designed to provide therapy on the basis of molecular signature rather than organ of origin, provided data that supported the U.S. Food and Drug Administration approval of two pan-Trk inhibitors — entrectinib and larotrectinib.3,4 NTRK fusions, as well as some splicing variants, have been reported in only a few hematologic malignancies including acute lymphoblastic leukemia, acute myeloid leukemia (AML), and histiocytic neoplasms.5-7 However, little is known about the contribution of NTRK point mutations to the pathogenesis of hematologic malignancies. Indeed, while patients treated with Trk inhibitors frequently develop secondary resistance mutations in the tyrosine kinase domain (TKD),2 early literature from solid tumors suggested that missense variants outside this domain might have little functional consequence.8
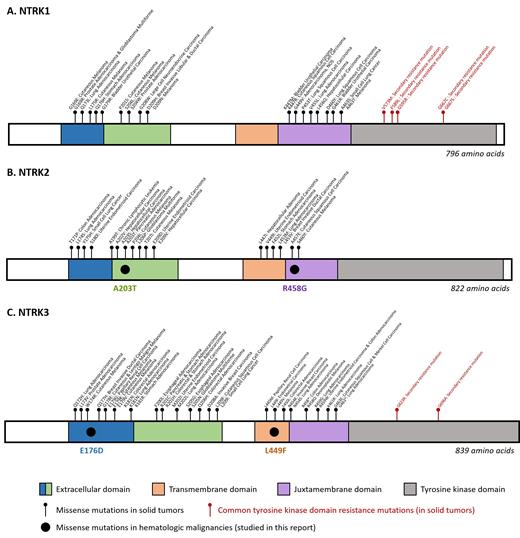
A recent study by Dr. Sunil K. Joshi and colleagues characterized NTRK point mutations in hematologic malignancies. Nine unique point mutations in NTRK2 and NTRK3 were identified by deep sequencing performed on 185 patient samples (96 AML, 51 acute lymphoblastic leukemia, and 38 myeloproliferative neoplasms). However, only four missense mutations (NTRK2A203T and NTRK3E176D found in the extracellular domains, NTRK3L449F found in the transmembrane domain, and NTRK2R458G in the juxtamembrane domain) were associated with cytokine-independent sustained proliferation of Ba/F3 cells — a measure of transforming ability. These four mutations amplified downstream signaling as demonstrated by increased phosphorylation of Trk, AKT, and ERK in transfected Ba/F3 and HEK293 cell lines. The study also shows that the mechanisms that activate the Trk signaling pathways are partially influenced by the mutation site. Mutations in the extracellular domain (NTRK2A203T and NTRK3E176D) and transmembrane domain (NTRK3L449F) were associated with increased Trk phosphorylation, transient elevation in Trk membrane expression, and Trk dimerization. However, a mutation in the juxtamembrane domain (NTRK2R458G) caused increased Trk phosphorylation but did not significantly alter Trk membrane expression or dimerization. Importantly, first-generation Trk inhibitors larotrectinib and entrectinib produced a dose-dependent decrease in Trk phosphorylation and an increase in apoptosis of mutant-transfected Ba/F3 cell lines.
In Brief
This impressive study by Dr. Joshi and colleagues sheds light on several key aspects and questions related to NTRK aberrations in hematologic malignancies. First, not all missense mutations result in activation of Trk function. This is an important caveat to all variant interpretation and may explain the early studies on solid tumor missense mutations that found no significant functional consequence of the mutations.8 Secondly, can solid tumors learn from hematologic malignancies about nonfusion alterations? Dr. Joshi and colleagues identified new domains in which pathogenic mutations can be found in the Trk proteins: extracellular domain, transmembrane domain, and juxtamembrane domain (Figure). Unlike the TKD mutations (NTRK1 p.G595R and NTRK3 p.G623R among others), which are secondarily acquired after treatment with first-generation Trk inhibitors (larotrectinib, entrectinib) that predominantly target NTRK fusions (Figure), these mutations seem to be primary and have variable function on Trk expression, dimerization, and/or internalization. Additionally, Dr. Joshi and colleagues show that these mutations are potentially targetable by first-generation Trk inhibitors. Therefore, although first-generation Trk inhibitors are ineffective against the secondary TKD mutations, they may be effective against some primary NTRK point mutations in other critical domains, thereby broadening the scope and applicability of these drugs, potentially in both hematologic and solid tumors. Third, should we routinely test for NTRK mutations in hematologic malignancies? NTRK point mutations were observed in approximately 5 percent of the study cohort, with the incidence of biologically characterized and pathogenic variants in 2.1 percent of cases, indicating that NTRK aberrations are rare in hematologic malignancies. However, NTRK fusions are even less common in solid tumors (0.3% of pan-cancer tumors),2 but due to the dramatic efficacy of targeted therapy for these fusions, are increasingly incorporated into solid tumor testing. Lastly, this study has illuminated a possible association between NPM1 and NTRK in AML. Interestingly, 40 percent of NTRK point mutations were observed in patients with NPM1-mutated AML patients, though only one of these NTRK mutations was functionally demonstrated to be likely pathogenic. However, these findings may indicate a potential association between NPM1 and NTRK mutations in AML. Studies in larger cohorts are necessary to characterize the frequency and potential prognostic significance of NTRK co-mutations in NPM1-mutated AML. In summary, the current study provides proof of concept that NTRK mutations drive hematologic malignancies and could serve as a launch pad for future studies in larger cohorts to determine the prognostic significance of NTRK aberrations in hematologic malignancies.
Structures of (A) NTRK1, (B) NTRK2, and (C) NTRK3 with the locations of the missense mutations in hematologic malignancies (studied in this report), missense mutations in solid tumors, and the common tyrosine kinase domain resistance mutations. (Adapted from Joshi SK, Qian K, Bisson WH, et al. Discovery and characterization of targetable NTRK point mutations in hematologic neoplasms. Blood. 2020;135:2159-2170, Suppl Figure 3.)
Structures of (A) NTRK1, (B) NTRK2, and (C) NTRK3 with the locations of the missense mutations in hematologic malignancies (studied in this report), missense mutations in solid tumors, and the common tyrosine kinase domain resistance mutations. (Adapted from Joshi SK, Qian K, Bisson WH, et al. Discovery and characterization of targetable NTRK point mutations in hematologic neoplasms. Blood. 2020;135:2159-2170, Suppl Figure 3.)
References
Competing Interests
Dr. Narayanan and Dr. Kim indicated no relevant conflicts of interest.