Myelodysplastic syndromes (MDS) are a group of clonal bone marrow failure disorders with variability in clinical phenotype, risk of transformation to acute myeloid leukemia (AML), treatment response, and ultimately, prognosis. The constellation of molecular aberrations affecting each individual patient has a major impact on clinical outcomes after treatment.1 The Revised International Prognostic Scoring System (IPSS-R) helps categorize patients with MDS into different risk categories2 and incorporates the extent of cytopenias at presentation, bone marrow blast percentage, and cytogenetic risk category3 ; however, molecular data are not currently included.
Nicknamed “the guardian of the genome,” tumor protein p53 (TP53) plays a role in carcinogenesis and independently predicts poor prognosis in MDS. Aberrations of the TP53 gene (in the form of mutations or deletions) notoriously associate with higher-risk disease, poorer response to therapies, and adverse outcomes.4,5 TP53-mutant MDS is considered a high-risk subtype, and outcomes are so poor that some authors have questioned whether intensive treatment and even allogeneic hematopoietic stem cell transplantation is futile in this group of patients.5 However, long-term survivors with TP53-mutant MDS are well described, suggesting that there may be nuance within this apparently genetically homogenous subtype.
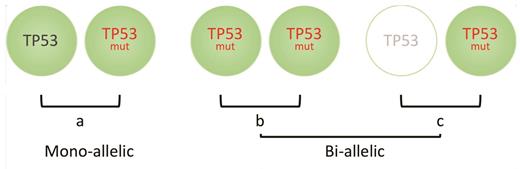
To this end, Dr. Elsa Bernard and colleagues have examined a very large clinical cohort of patients with MDS and were able to demonstrate that not all TP53 aberrations have equivalent impact on survival.6 Extended genetic profiling was performed on a total of 4,444 patients and consisted of conventional cytogenetic analysis, targeted next-generation sequencing of genes known to be recurrently mutated in MDS, along with genome-wide copy number probes detecting the presence of allelic imbalances. Four TP53-mutant subgroups were defined; 33 percent of patients had monoallelic mutations of TP53 (a single gene mutation), 24 percent had more than one TP53 mutation with no deletion or copy-neutral loss of heterozygosity (cn-LOH) affecting the TP53 locus, 22 percent of patients had mutations and deletion of the TP53 locus, and 21 percent had mutations and concomitant cn-LOH (the latter two groups with no effective TP53). These groups were then able to be categorized into two broader groups depending on whether one TP53 allele was affected (monoallelic) with one residual wildtype TP53 allele, versus both alleles affected with no residual TP53 (biallelic or multi-hit; Figure). Dr. Bernard and colleagues demonstrated differences in prognostic features between the two groups including clinical characteristics, karyotype, and survival.
(a) Patients with myelodysplastic syndrome (MDS) who have mono-alleleic mutations in TP53 have similar outcomes to TP53-unmutated MDS. Conversely, bi-allelic loss caused by either (b) multiple mutations of TP53, copy-neutral loss of heterozygosity or (c) mono-allelic mutation of TP53 with deletion of the other TP53 allele, will result in no wildtype TP53 expression, and these groups are associated with inferior clinical outcomes.
(a) Patients with myelodysplastic syndrome (MDS) who have mono-alleleic mutations in TP53 have similar outcomes to TP53-unmutated MDS. Conversely, bi-allelic loss caused by either (b) multiple mutations of TP53, copy-neutral loss of heterozygosity or (c) mono-allelic mutation of TP53 with deletion of the other TP53 allele, will result in no wildtype TP53 expression, and these groups are associated with inferior clinical outcomes.
Patients with biallelic hits to TP53 (multi-hit), regardless of the nature of the “hit” (mutation or deletion), were associated with established high-risk features such as complex karyotype.3 Higher number of chromosomal aberrations were seen per patient, with 91 percent of multi-hit patients having a complex karyotype in comparison to 13 percent of monoallelic patients, suggesting that the presence of residual TP53 activity is essential in maintaining the stability of chromosomes, which is also supported by data from Dr. Guillermo Montalban-Bravo and colleagues.7 In comparison, monoallelic patients had less severe cytopenias and lower bone marrow blast percentages, similar to levels seen in wild-type TP53, contributing to lower-risk IPSS-R. Notably, 10 percent of multi-hit patients were classified as “very good” to “intermediate” risk using the IPSS-R, highlighting a subset of patients with higher-risk disease undetected using conventional risk scoring methods. Median overall survival (OS) was significantly different between the two groups at 8.7 months for the multi-hit versus 3.5 years for the monoallelic group.
As previously reported, TP53 variant allele frequency (VAF) was noted to have prognostic significance associated with complex karyotype and poor treatment responses.7,8 High VAF was strongly correlated with biallelic targeting, which may explain the poor outcomes. However, a proportion of patients with cn-LOH affecting the TP53 locus had VAFs less than 50 percent, demonstrating that VAF estimates alone are not sufficient in evaluating allelic state. Pattern of co-mutations were also notably different between monoallelic and multi-hit groups, with no driver mutations seen in multi-hit TP53 whilst up to 90 percent of monoallelic patients had at least one other driver mutation. This pattern of higher VAF, poor disease characteristics, and lack of associated driver mutations with multi-hit TP53 suggests early aberration of TP53 function in the clonal evolution with these patients. In contrast, monoallelic TP53 states were frequently associated with lower VAFs consistent with subclones and co-occurred with a broad range of gene mutations such as TET2, SF3B1, ASXL1, RUNX1, and JAK2, amongst others.
Multi-hit TP53 identifies a very high-risk group of patients independently of the IPSS-R and molecular profile. Emerging target therapies may play a role in improving outcomes in patients with mutant TP53. APR-246, a methylated derivative of PRIMA-1, restores the transcriptional transactivation function of mutant TP53, thereby inducing apoptosis of the affected cell. Preclinical trials show a synergistic effect of APR-246 with 5-azacitidine in TP53-mutated MDS.9 A phase III multicenter randomized trial is underway comparing this combination in patients with MDS with the standard of care, 5-azacitidine alone (clinicaltrials.gov identifier, NCT03745716). Challenges remain, however, as targeted therapies rely on the presence of a mutant TP53 copy and may not be effective where there is complete TP53 deletion.
Personalized genomic medicine benefits from a precise understanding of the clinical and functional impact of specific genetic aberrations. This article illuminates the importance of determining TP53 mutational status in patients with MDS at diagnosis, but also extends these findings to demonstrate the importance of mono- versus bi-allelic loss of TP53 function and allele dosage. Importantly, it can identify a subgroup of monoallelic TP53-mutant MDS that has similar clinical outcomes to non-TP53 mutated MDS. This work will have a substantial clinical impact and therefore deserves this selection as one of the Year’s Best.
References
Competing Interests
Dr. Jivan and Dr. Lane indicated no relevant conflicts of interest.