Small non-coding RNA, including microRNA (miRNA), regulate gene expression through either degradation of messenger RNA or suppression of translation. The capacity of miRNA to regulate lineage commitment, differentiation, and self-renewal of hematopoietic stem and progenitor cells (HSPC) is becoming increasingly recognized.1 Studies of their physiologic impact are often hindered by the abundance of mRNA target transcripts and the diverse functional effects that aberrant expression of these regulators produce. Understanding miRNA regulation in HSPCs is further complicated by the fact that the majority of miRNAs are normally expressed at very low copy numbers. Existing studies of miRNA in HSPCs, therefore, have often taken a candidate approach in which a small number of miRNAs and their targets are analyzed using in vitro model systems. Interpretation of these studies is frequently compromised by the absence of in vivo functional validation.
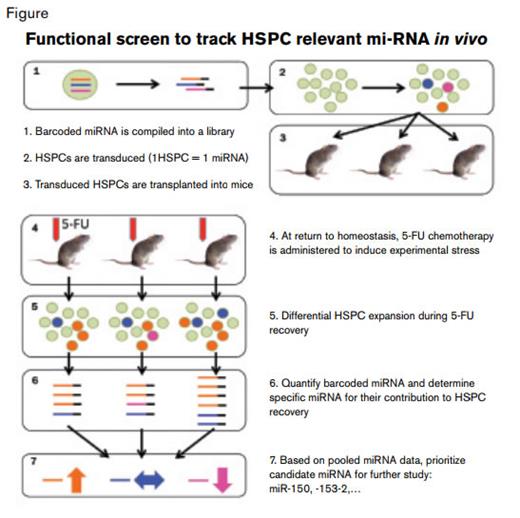
In the current paper, Adams and colleagues from Yale University School of Medicine use an elegant in vivo screening method that lets cell function guide discovery of miRNAs involved in HSPC regeneration. Their functional screen hinged on the transduction of hematopoietic cells with a pool of retroviral vectors encoding a set of HSPC-relevant miRNA. The flanking sequence of each vector was tagged with a genomic sequence that served as a unique barcode for recovery and identification of cells expressing a particular transgene. Cells, transduced at limiting efficiency to favor single-copy integration (i.e., 1 miRNA per HSPC), were then transplanted into lethally irradiated recipients. Once stable hematopoietic function had been regained after transplantation, the animals were challenged with a chemotherapy agent, 5-fluorouracil (5-FU). The hypothesis being tested posited that during the subsequent recovery from 5-FU-induced stress, stem cells expressing a particular provirus-encoded miRNA would be predominant in the repopulated marrow, while those expressing other miRNAs would be relatively underrepresented. Because each animal provides a screen for several HSPC clones, each of which carries a unique barcoded miRNA, a moderately sized library of transcripts (135) could be evaluated in a relatively small number of mice (15) by correlating changes across three independent transplant experiments.
Based on specific miRNA representation in animals during recovery from 5-FU, the authors earmarked a group of miRNAs for further study. Micro RNAs that were overrepresented in transplanted animals contributed to hematopoeitic recovery after injury, while those that were underrepresented negatively impacted this process. Focusing on the latter class of suppressive miRNAs, miR-153-2, miR-150, miR-301, and miR-652 were selected for further characterization. Through a series of competitive repopulation studies of wild-type marrow and HSPCs transduced to overexpress candidate miRNAs, they identified miR-153-2, miR-150, and miR-652 as inhibitors of myeloid and platelet recovery, with miR-150 overexpression resulting in the greatest suppressive effect. The authors then painstakingly validated the results using a number of different in vivo and in vitro experimental approaches. They demonstrated decreased colony-forming capacity and increased apoptosis after 5-FU treatment in cells that overexpressed miR-150, which was partially phenocopied in a model of a well-established target transcript of miR-150, c-myb. Collectively these experiments convincingly implicate a specific miRNA (i.e., miR-150) in suppression of bone marrow recovery after chemotherapy-mediated injury.
In Brief
This work is notable on at least three levels. First, inspired by genetic approaches in other model systems, the functional in vivo screen for miRNA circumvents many of the difficulties inherent in examining miRNA-mediated stem cell regulation. It allows investigators to sift through the large number of potential targets that emerge from conventional in silico screens and focus their efforts on those that are the most likely to be relevant to the phenotype of interest. This method offers gains in efficiency and stringency compared with broad observational studies. Second, the screen can be readily adapted to identify miRNAs that are involved in regulation of hematopoiesis in a wide variety of benign and malignant conditions. Third, even when viewed as a proof-of-concept report, the identification of miR-150 as a negative regulator of hematopoietic recovery after chemotherapy-mediated marrow injury is significant. This finding, along with emerging data on the role of miR-150 in leukemogenesis, provides the rationale for additional study.
References
Competing Interests
Mr. Hornick and Dr. Kurre indicated no relevant conflicts of interest.