Long ago, in the guest bedroom of my grandparents’ summer cottage, there stood a battered 19th century French Provincial oak dresser so massive that if Atlas himself had been asked to lift it, he might have shrugged and walked away. Faux-weathered wood with its rustic, vintage connotations is all the rage in the decorative arts world these days, but that old dresser came by its shabbily genteel appearance honestly, through a lifetime of heavy use – every nick and dent silently bearing witness to a history of laborious relocations and close encounters with chairs, each chip and scrape memorializing the trajectory of an impatiently shoved vacuum cleaner or an errant John Deere toy.
Data from the Genome Institute at Washington University in St. Louis indicate that just as a wellloved piece of furniture accumulates wear and tear over the years, the DNA of hematopoietic stem and progenitor cells (HSPCs) endures a similar lifelong steady beating. Whole-genome sequencing (WGS) of HSPCs from umbilical cord blood, septuagenarian marrow, and healthy volunteers from every age cohort in between demonstrates age-dependent acquisition of somatic mutations, most commonly C>T/A>G transitions resulting from random deamination of methylated cytosine nucleotides. Other classes of mutations were shown to emerge with aging at the expected rate for DNA replication infidelity.
When, on a very bad day, one of these mutations arises in a particularly unfortunate location, malignant transformation to acute myeloid leukemia (AML) or another neoplasm can result. The cell’s genomic signature, at that inauspicious moment, captures its idiosyncratic history of nucleic acid trauma. The characteristic malignant behavior of the neoplastic clone might be driven by a FLT3 or DNMT3A mutation, but all of the other genomic scars – private mutations located in genes encoding proteins consequential only in, say, the retina or the ovary, or sited in meaningless intronic space far from the coding and regulatory bits of the genome – are still there, in their hundreds, bearing a silent story the way a manatee’s scarred hide tells tales of close encounters with long-gone ship propellers.
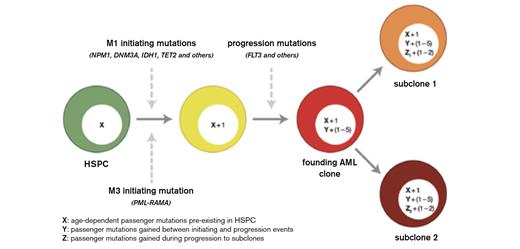
Integrated Model for the Origin of Driver and Passenger Mutations During AML Evolution. Hematopoietic stem/progenitor cells (HSPCs, shown in green) accumulate random, benign background mutations as a function of age. “X” represents these background mutations in the HSPCs and may range from ~100 to 1,000 events, depending on age. The initiating mutations are different for M1 AML cases and M3 AML cases. Because the initiating event provides an advantage for the affected cell, clonal expansion ensues, so that all of the preexisting mutations in that cell are “captured” by cloning. Each cooperating mutation gives the expanding clone an additional advantage; our data suggest that one to five events contribute to progression in most cases of AML. Each cooperating mutation is expected to capture all mutations that occurred between the initiating event and the progression event (designated as “Y” in the yellow cell). Although this number is unknown, the analysis of clonal progression of secondary AML suggests that each cooperating mutation may capture dozens to hundreds of mutations. The “founding” AML clone is designated in red. Subclones arise from the founding AML clone by acquiring a small number of additional mutations that confer an advantage to that cell, along with any additional background mutations that may have occurred in the interim (represented as “Z”).Welch JS et al. Cell. 2012;150:264-278. Reprinted with permission of Elsevier.
Integrated Model for the Origin of Driver and Passenger Mutations During AML Evolution. Hematopoietic stem/progenitor cells (HSPCs, shown in green) accumulate random, benign background mutations as a function of age. “X” represents these background mutations in the HSPCs and may range from ~100 to 1,000 events, depending on age. The initiating mutations are different for M1 AML cases and M3 AML cases. Because the initiating event provides an advantage for the affected cell, clonal expansion ensues, so that all of the preexisting mutations in that cell are “captured” by cloning. Each cooperating mutation gives the expanding clone an additional advantage; our data suggest that one to five events contribute to progression in most cases of AML. Each cooperating mutation is expected to capture all mutations that occurred between the initiating event and the progression event (designated as “Y” in the yellow cell). Although this number is unknown, the analysis of clonal progression of secondary AML suggests that each cooperating mutation may capture dozens to hundreds of mutations. The “founding” AML clone is designated in red. Subclones arise from the founding AML clone by acquiring a small number of additional mutations that confer an advantage to that cell, along with any additional background mutations that may have occurred in the interim (represented as “Z”).Welch JS et al. Cell. 2012;150:264-278. Reprinted with permission of Elsevier.
John Welch, Tim Ley, Dan Link, and their Wash U. colleagues further explored the origin and clonal evolution of AML by WGS of 12 patients with normal karyotype AML M1, as well as 12 patients with acute promyelocytic leukemia (APL) who have a clear disease-initiating molecular event, PML-RARA translocation. An average of 10 to 11 mutations with translational consequences was present in the AML cases, but most mutations were not recurrent (Figure). The differences between the M1 and APL results are also interesting and could have filled another paper; briefly, mutations more common in M1 AML than APL (e.g., NPM1, IDH1, DNMT3A – 13 recurrently mutated genes were found only in M1 and not APL) might represent initiating events, while the nine recurrent mutations (e.g., FLT3) common to both subtypes are likely to be cooperators. This hypothesis is supported by mouse models in which FLT3 mutations are not enough to initiate leukemia.
In Brief
As with all good science, this new work raises as many questions as it answers. Are some of the mutations that at first blush appear to be inconsequential “passengers” actually important cooperative events? The 15 detected recurrently mutated non-genic regions, for instance, are attractive candidates for exploration. What determines which subclones contribute to relapse? And, mechanistically, how do newly described recurrent mutations contribute to leukemogenesis, such as those in the cohesin complex? In his E. Donnall Thomas Lecture at the 2012 ASH Annual Meeting, Dr. Ley stated that all of the AML-associated mutations present in at least 5 percent of patients have probably already been described. But while the gateway to the garden of discovery for common recurrent mutations in AML is rapidly closing, there are plenty of new wilds awaiting energetic explorers.
Competing Interests
Dr. Steensma indicated no relevant conflicts of interest.