The phospholipid composition of the plasma membranes of most eukaryotic cells is composed primarily of phosphatidylcholine (PC); sphingomyelin; and two aminophospholipids, phosphatidylserine (PS) and phosphatidylethanolamine (PE). PC and sphingomyelin are asymmetrically distributed to the outer leaflet of the membrane, whereas PS and PE are located primarily in the inner leaflet. Disruption of the lipid asymmetry and movement of PS to the outer leaflet results in phagocytosis by macrophages of cells undergoing apoptosis. Additionally, PS exposure on the platelet surface promotes assembly of coagulation complexes, which is necessary for normal hemostasis.
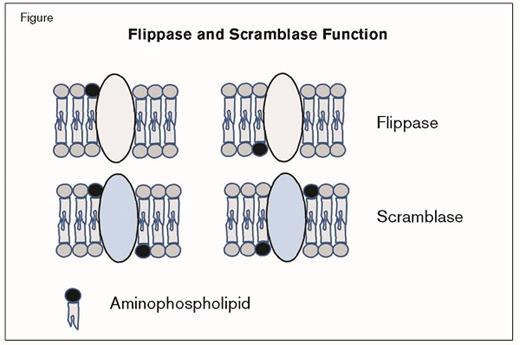
Phospholipid membrane asymmetry is regulated by three types of enzymatic activity.1 Flippases are ATP-dependent translocases that transport aminophospholipids from the outer leaflet to the inner leaflet (Figure). Conversely, floppases transport lipids from the inner leaflet to the outer leaflet. Scramblases are ATP-independent translocases that move aminophospholipids in both directions (Figure). Although TMEM16F and Xkr8 have been identified as scramblases involved in apoptosis and platelet function, respectively,2,3 the enzyme(s) responsible for flippase activity are not well characterized. Leading candidates for flippase activity are members of the type 4 subfamily of P-type adenosine triphosphatases (P4-ATPases), including ATP11C.1
Dr. Katsumori Segawa and colleagues in the laboratory of Shigekazu Nagata at Kyoto University used a gene-trapping strategy4 to identify and characterize flippase function in mammalian cells. A retroviral vector was used to introduce inactivating insertions in the haploid KBM7 human cell line. The advantage of haploid cells over diploid cells is that inactivation of one allele rather than two alleles is required to induce complete loss of function. An additional advantage of the gene-trapping method used by Dr. Segawa and colleagues is that the integrated DNA sequences provided a tag that allowed the investigators to readily identify the disrupted gene. Loss of flippase function in KBM7 cells was assayed by measuring cellular uptake of the fluorescent dye, 7-Nitro-2-1,3-benzoxadiazol-4-yl–tagged PS (NBD-PS), a process that requires flippase activity. Mutagenized cells with low flippase activity were identified by flow cytometry and isolated by sorting. Gene trap insertion sites in the isolated low-flippase cells were identified in two genes, ATP11C and CDC50A, the latter of which had previously been identified as the gene encoding the β subunit of ATP11C. Low flippase expression in cloned, gene-trapped ATP11C-deficient or CDC50A-deficient KBM7 cell lines was rescued by introducing the active ATP11C or CDC50 genes, respectively, using retroviral vectors, confirming the flippase activity associated with those genes.
To study flippase function during apoptosis, murine W3 cells, which undergo apoptosis following treatment with Fas ligand (FasL) were employed. The ATP11C gene in W3 cells was mutated using a recently described site-specific mutagenesis method that takes advantage of the CRISPR/Cas enzyme system.5 W3 cell lines that contained ATP11C truncations and loss of flippase activity were identified. Treatment of wild-type W3 cells with FasL resulted in the proteolysis of ATP11C, which was blocked by the caspase inhibitor QVD-OPh. An ATP11C gene, designated CasR, was constructed containing mutations at three putative caspase recognition sites and was introduced into ATP11C-deficient W3 cells. ATP11C in the CasR cell line was no longer subject to caspase-cleavage, but retained flippase activity. Importantly, the CasR cell line did not expose PS upon FasL treatment. ATP11C-deficient W3 cells transformed with the human ATP11C gene were phagocytosed by macrophages in an in vitro system, but CasR-expressing cells were not. These results indicate that cleavage and inactivation of ATP11C by caspase are required to inactivate flippase function and allow PS exposure on the cell surface.
The CDC50A gene in W3 cells also was mutated using the CRISPR/Cas system. Cell lines containingCDC50A truncations displayed loss of flippase function, exposed PS on the cell surface, and could not support the expression of ATP11C on the plasma membrane. These functional defects were rescued by transformation with the wild-type CDC50A gene. Living (non-apoptotic) CDC50A-deficient cells, but not CDC50A-rescued cells, were engulfed by macrophages invitro. Engulfment of living CDC50A-deficient KBM7 cells by macrophages was also observed, and engulfment was blocked by D89E MFG-E8, a protein that binds PS.
In Brief
The results of this study demonstrate that ATP11C is a PS flippase and that CDC50A is a functional subunit of ATP11C. The finding that macrophages engulf CDC50A-deficient cells indicates that live, non-apoptotic cells can be engulfed if PS is exposed. The possible clinical relevance of this finding is underscored by the observation that phagocytosis of living, healthy neurons may play a role in the pathophysiology of neurodegenerative disorders, and, conversely, evasion of phagocytosis by cancer cells may contribute to malignancy.6 The results of this study also provide a possible explanation for the observation that B-cell maturation in ATP11C-defective mice is abnormal. If ATP11C were the only P4-ATPase expressed in early B-cell development, ATP11C-deficient cells would expose PS and be cleared by macrophages.
References
Competing Interests
Dr. Lollar indicated no relevant conflicts of interest.