More than 250 mutations have been identified in acute myeloid leukemia (AML) that are seen recurrently in more than one patient. Some mutations have been classified as initiating mutations that occur early, such as NPM1, DNMT3A, IDH1, and TET2, and some mutations are cooperating mutations, such as FLT3, which activates signaling pathways. Driver mutations provide a survival or proliferative advantage to the cell that promotes the development of AML.1,2 There is also acquisition of additional subclonal mutations over time and after treatment,3 a process known as “clonal evolution.” Whole genome or exome sequencing of 200 genomes from patients with AML for The Cancer Genome Atlas (TCGA) revealed on average 13 gene mutations per genome, and 23 genes were found to be significantly mutated, with a higher-than-background mutation rate, likely to represent driver mutations. Several other studies have been published since these earlier reports, including one showing that mutations in secondary AML arising from myelodysplastic syndrome (MDS) are largely in the same genes as primary AML.4 However, it has been elusive to define how this genomic knowledge will enhance our practice.
Chromosomal analysis by standard karyotyping and/or fluorescence in situ hybridization (FISH) permits subdivision of patients into favorable, intermediate, or unfavorable-risk groups. There have been multiple attempts to identify biomarkers that can accurately predict the durability of response in AML, and to better subdivide intermediate-risk patients. One such marker for patients in complete remission (CR), classically defined as less than 5 percent bone marrow (BM) blasts, is minimal residual disease (MRD) by multicolor flow cytometry.5,6 Patients who attain CR but still have MRD of any amount identifiable by flow cytometry have a worse progression-free and overall survival.
Dr. Jeffrey Klco and collaborators, report that detection of mutations that persist on analysis of the day-30 BM is also predictive of shorter event free and overall survival (EFS and OS) in patients who achieve CR after initial therapy. 68 of the TCGA patients received induction chemotherapy with anthracycline (daunorubicin or idarubicin) and continuous infusion cytarabine at either 100 or 200 mg/m2 /day, and three additional similarly-treated patients were added to this group. Of these 71 patients, 34 were refractory, 12 relapsed between six and 12 months, and 25 survived at least 12 months without an allogeneic transplant. For these three groups, there were no differences in the number of detectable subclones, the number of total genomic variants, or coding mutations.
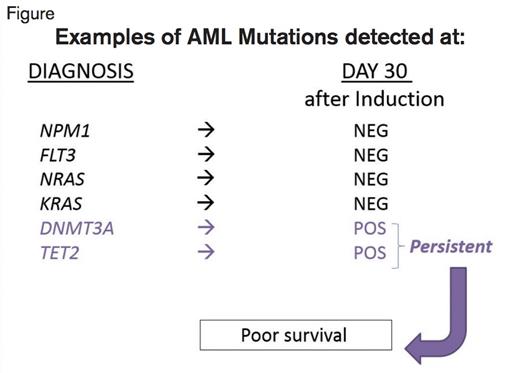
Examples of Results of Testing for Mutations at Diagnosis and on the Bone Marrow Obtained About Day 30 After the Start of Induction Chemotherapy. At diagnosis, the listed mutations are examples of those seen in some of the patients. The column “Day 30” illustrates whether those mutations were detected (POS, positive) or not detected (NEG, negative) at the time the bone marrow was obtained, at about day 30. The patients with mutations that were still present at day 30 exhibited worse EFS and OS compared with those that cleared the mutations.
Examples of Results of Testing for Mutations at Diagnosis and on the Bone Marrow Obtained About Day 30 After the Start of Induction Chemotherapy. At diagnosis, the listed mutations are examples of those seen in some of the patients. The column “Day 30” illustrates whether those mutations were detected (POS, positive) or not detected (NEG, negative) at the time the bone marrow was obtained, at about day 30. The patients with mutations that were still present at day 30 exhibited worse EFS and OS compared with those that cleared the mutations.
The study group comprised 25 patients with sufficient DNA from paraffin-embedded samples at pre-treatment and 30-days post treatment, and an additional 25 patients for whom they had cryopreserved samples at diagnosis, first remission, or at relapse. All 50 patients achieved CR with less than 5 percent BM blasts at a median of 34 days after the start of induction. Certain mutations did not clear by day 30. For example, of the 16 cases with DNMT3A mutations, only three cleared by day 30. Similarly, mutations in TET2 often persisted in remission. Mutations in FLT3, NRAS, and KRAS were cleared below the level of 5 percent of the cells with the mutation (< 2.5% variant allele frequency for a heterozygous mutation), as this level is believed to be reliable and not due to artifact of the methodology. All 18 cases with an NPM1 mutation cleared it by day 30. There were 24 patients who had at least one mutation detectable at that level or higher, and these patients exhibited a reduced median EFS of only six months compared with 17.9 months for the 26 patients who cleared the mutations. The respective median OS rates for these two groups were 10.5 months versus 42.2 months. Moreover, for patients with intermediate-risk karyotype, the same reduction of median EFS (8.8 vs. 25.6 months) and OS (19.3 vs. 46.8 months) was observed if mutations were still detected on day 30.
In Brief
There has been considerable difficulty in finding utility in the identification of what now amounts to hundreds of mutations in AML. Investigators continue to struggle to find a practical application of next generation sequencing in terms of modifying treatment, with few exceptions (e.g., FLT3). As there are already alternative methods to identify MRD with rapid turnaround times (~1 day) and higher sensitivity, such as multicolor flow cytometry (sensitivity at least 0.1%) and FISH (sensitivity in the range of 1% to 7%), it remains to be seen under which circumstances the mutation testing described by this study would be superior to current methods. There may be selected circumstances (for example, patients with normal karyotype who have well-defined mutations associated with prognosis) for which such monitoring may be helpful, especially if a rapid method could be developed that would enhance the reliability of detection of mutations present in less than 0.1% of the cell population. Only the future will tell.
References
Competing Interests
Dr. Becker has received a speaker's honorarium as well as travel funds from Invivoscribe Technologies/Genection.