Since the invention of flow cytometry by Dr. Leonard Herzenberg and colleagues in the 1960s,1 the technology available to identify and quantify cells on a single-cell basis has progressed to the extent that identifying many cell types simultaneously in complex, heterogeneous tissues such as blood and bone marrow, and measuring these cells individually for multiple physiological parameters is now possible. A major recent advance in cytometric methods has been the development of mass cytometry, which replaces the well-established labeling of antibodies with fluorescent dyes, with the use of metal-labeled “mass tags,” which can be identified by a mass spectroscopic readout.2 The metal tags currently available commercially are predominantly lanthanides, from 141-Pr to 176-Yb, which are extremely rare in biological tissue and therefore have no intrinsic cell-derived background signal. Routine experiments are now performed in which 30 or more distinct mass tag–labeled antibodies are applied to the experimental samples, thereby allowing researchers to quantify at least 30 molecules at the single-cell level.
Even prior to the development of mass cytometry, the need for monitoring features of cancer cell physiology at the single-cell level has been evident. The identification of hyperactivating mutations in genes encoding signaling molecules, such as BCR-ABL1, FLT3, and JAK2, has made monitoring the activities of these and downstream signal transducers a valuable proxy for an active neoplasm. Cancer-signaling phenotypes can be identified based on abnormal signal transduction and studied to predict features of the cancer such as prognosis3 and sensitivity to drug treatments.4 Antibodies that recognize individual phosphorylated sites on signaling molecules, such as cytokine receptors and kinases (as well as their downstream effectors), can be used to measure the activities of intracellular signaling pathways based on the quantitative intensity of antibody labeling. In a pioneering study, single-cell flow cytometric analysis was used to characterize altered signaling networks in acute myeloid leukemia (AML) patient cells, demonstrating that specific signaling signatures could be correlated with prognosis and response to chemotherapy.5 Subsequent studies have applied similar approaches to identify pathophysiologic signaling responses in other myeloid neoplasms as well as lymphoid malignancies.6-9
The advent of mass cytometry allows researchers to account for complex features that would be much more difficult to study by fluorescent flow cytometry because of the limited number of nonconflicting fluorophore channels per experiment. With mass cytometry, features of intratumor heterogeneity, such as the identification of stem cells and derivative cells, can be studied, which may offer crucial distinctions in stem cell–propagated neoplasms. Interactions between tumor and non-tumor cells can be studied in patient samples.7 Such approaches enable the characterization of dysregulated signaling networks in cell subsets spanning the entire hematopoietic continuum – a focus of our own research on myeloproliferative neoplasms.10
The multidimensional data derived from mass cytometry experiments was originally described in a study illustrating the signaling behavior of healthy human bone marrow mononuclear cells.4 In this study, 33 immunophenotypic markers were utilized to identify 29 cell subsets. In these populations, 18 antibodies specific for intracellular signaling modifications (mainly phosphorylation of signaling effectors) were assayed in response to 15 perturbations, including exposure to cytokines and targeted inhibitors such as dasatinib. This study now serves as an important baseline reference to which signaling responses in disease states can be compared.
The analysis of multidimensional data derived from mass cytometry has been enabled by the development of several bioinformatics tools. The SPADE (Spanning tree Progression of Density normalized Events) clustering algorithm groups cells stochastically based on shared immunophenotypic marker labeling, and can be used to compare cell signaling in selected cell populations across multiple experimental samples.4,11 The more recently developed tool viSNE (Visualization of t-distributed Stochastic Neighbor Embedding algorithm) has the advantages of reproducibility and visualization of signaling in individual cells (as opposed to grouped cell clusters).12 Both SPADE and viSNE are available through the online analysis platform Cytobank (www.cytobank.org). Both analytic tools have been used in recent studies of hematologic cancers.12-14
Another recent study has added two new analytic algorithms, PhenoGraph and SARA (Statistical Analysis of Response Amplitude), to the mass cytometry analyst’s toolkit.3 These tools allowed the generation of a ‘multidimensional immunophenotypes-by-signaling-phenotypes’ matrix, which was used to identify cell surface and signaling phenotypes associated with poor prognosis across multiple molecular subtypes of AML.3 The signaling phenotypes enabled a prognostic predictive value that could not be achieved from cell surface immunophenotypes of the AML blasts alone. The power of newly developed analytic tools, coupled with the multidimensional quantitative data collection enabled through mass cytometry, will further our understanding of the development and progression of hematologic malignancies.
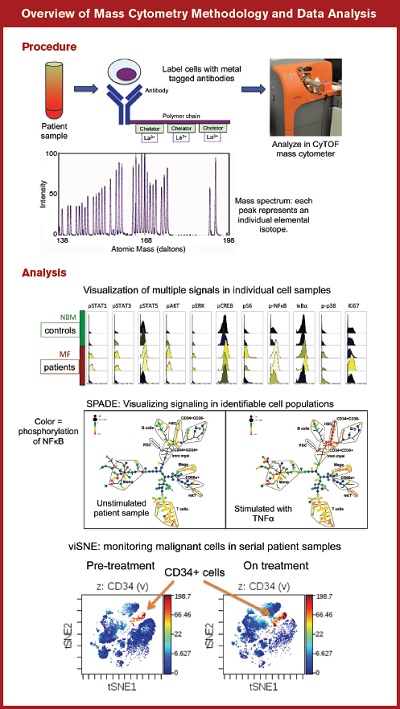
Overview of Mass Cytometry Methodology and Data Analysis.Procedure: Top, cells isolated from patient or control samples are labeled with metal tagged antibodies, whose atomic masses are read by the CyTOF mass cytometer. Below, a spectrum of atomic masses of lanthanide metals used to label antibodies for mass cytometry.Analysis: Top, multiple signaling molecules can be measured simultaneously and compared in patient versus control sample cell populations. Middle, SPADE (Spanning tree Progression of Density normalized Events) analysis illustrates signaling across multiple cell populations basally and in response to stimulation with TNFα (tumor necrosis factor-α). Bottom, viSNE (Visualization of t-distributed Stochastic Neighbor Embedding algorithm) plots identifying the persistence of CD34+ stem/progenitor cells in the peripheral blood of a myelofibrosis patient prior to (left) and on treatment (right) with ruxolitinib.
Overview of Mass Cytometry Methodology and Data Analysis.Procedure: Top, cells isolated from patient or control samples are labeled with metal tagged antibodies, whose atomic masses are read by the CyTOF mass cytometer. Below, a spectrum of atomic masses of lanthanide metals used to label antibodies for mass cytometry.Analysis: Top, multiple signaling molecules can be measured simultaneously and compared in patient versus control sample cell populations. Middle, SPADE (Spanning tree Progression of Density normalized Events) analysis illustrates signaling across multiple cell populations basally and in response to stimulation with TNFα (tumor necrosis factor-α). Bottom, viSNE (Visualization of t-distributed Stochastic Neighbor Embedding algorithm) plots identifying the persistence of CD34+ stem/progenitor cells in the peripheral blood of a myelofibrosis patient prior to (left) and on treatment (right) with ruxolitinib.
A particularly valuable application of mass cytometry and its corresponding analytic tools will be the analysis of disease evolution in serial patient samples and response to specific therapies. Recent studies have utilized viSNE to identify minimal residual disease populations during treatment and relapse of small numbers of patients with AML and ALL.12,14 Expanding these studies to larger patient cohorts may enable characterization of disease-resistant cell populations, identification of cell signaling phenotypes that predict risk of relapse, and ultimately, discovery of potential therapeutic vulnerabilities of malignant cells that resist previously established treatment.
Figure 1. Overview of mass cytometry methodology and data analysisProcedure: Top, cells isolated from patient or control samples are labeled with metal tagged antibodies, whose atomic masses are read by the CyTOF mass cytometer. Below, a spectrum of atomic masses of lanthanide metals used to label antibodies for mass cytometry.Analysis: Top, multiple signaling molecules can be measured simultaneously and compared in patient versus control sample cell populations. Middle, SPADE (Spanning tree Progression of Density normalized Events) analysis illustrates signaling across multiple cell populations basally and in response to stimulation with TNFα (tumor necrosis factor-α). Bottom, viSNE (Visualization of t-distributed Stochastic Neighbor Embedding algorithm) plots identifying the persistence of CD34+ stem/progenitor cells in the peripheral blood of a myelofibrosis patient prior to (left) and on treatment (right) with ruxolitinib.
References
Competing Interests
Dr. Oh has received honoraria from Fluidigm Corporation. Dr. Fisher has no relevant conflicts of interest.