More than 15 years ago, the Human Genome Project provided a comprehensive sequence of our genome’s gene-containing regions. We now have the ability to directly edit DNA in a systematic manner using a variety of techniques, allowing the comprehensive study of the function of all genes.
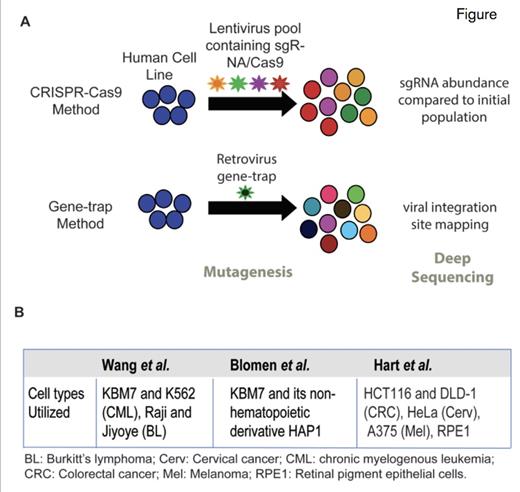
Functional Screens to Identify Essential Genes in Human Cells. (A) Two approaches taken by Dr. Tim Wang and coauthors to identify genes essential for human cell survival. One approach utilized CRISPR-mediated gene deletion using pools of small-guide RNAs (sgRNAs) targeting all protein-coding genes (top). They simultaneously utilized retroviral-mediated insertion of gene traps into the coding sequence of all protein-coding genes (bottom). (B) Table depicting the cell types used in similar functional genomic screens to identify essential genes in three recent studies.
Functional Screens to Identify Essential Genes in Human Cells. (A) Two approaches taken by Dr. Tim Wang and coauthors to identify genes essential for human cell survival. One approach utilized CRISPR-mediated gene deletion using pools of small-guide RNAs (sgRNAs) targeting all protein-coding genes (top). They simultaneously utilized retroviral-mediated insertion of gene traps into the coding sequence of all protein-coding genes (bottom). (B) Table depicting the cell types used in similar functional genomic screens to identify essential genes in three recent studies.
Three independent studies have recently furthered our understanding of those genes that are indispensable for the viability of human cells.1-3 One of those studies, by Dr. Tim Wang and colleagues, utilized a near-haploid human chronic myelogenous leukemia (CML) cell line called KBM7 (karyotype: 25, XY, +8, Ph+) to disrupt DNA coding sequences throughout the genome and uncover which genes are essential to the survival of these cells.1 Using a CRISPR/Cas9–based gene-editing strategy with pools of lentiviruses containing small-guide RNA (sgRNA), they targeted every protein-coding gene in the genome (Figure A). Comparison of sgRNA abundance between the initial infected cell population and 14 population doublings then identified those sgRNAs and the corresponding genes, which were depleted in the final population. In an orthogonal approach, they also utilized retroviral gene-trap mutagenesis to disrupt the coding sequence of all protein-coding genes. Integration of a gene-trap in the sense orientation within the coding region of a gene results in gene inactivation while integration in the antisense has no effect. The proportion of gene-trap insertions in the activating versus inactivating orientation for each gene in the genome thereby allows for deduction of the genes that have an essential function. A similar methodology of gene-trap insertional mutagenesis in KBM7 cells and an additional cell line derived from KBM7 cells, was also published by Dr. Vincent A. Blomen and colleagues in the same issue of Science magazine.2 Finally, Dr. Traver Hart and colleagues utilized a genome-wide CRISPR negative-selection screen across five human cancer cell lines to identify core fitness genes (Figure B).3
The results from each of the three mentioned studies were enlightening and highly analogous in their findings. First, all three made the surprising finding that approximately 2,000 genes (nearly 10% of the genes in the human genome) are essential. The majority of essential genes have similar attributes that reflect their critical roles: they are often conserved amongst species, evolve slowly, rarely have paralogs, are highly expressed, tend to be rich in protein-protein interactions, and are rarely affected by nonsynonymous mutations. More surprisingly, roughly 300 of the essential genes in our genome have no previously studied function. The discovery of these newly identified essential genes was made possible through the systemic use of techniques to directly mutagenize DNA, identifying fourfold to fivefold more essential genes compared with prior similar efforts using RNA interference.
In Brief
While the studies by Dr. Wang and Dr. Hart and their colleagues begin to define genes that appear to be essential in the context of specific cancer oncogenes (such as BCR-ABL1 and KRAS G13D), these represent only the beginning of such efforts.1,3 For example, the study by Dr. Blomen and colleagues revealed that those genes that are nonessential on their own may actually result in cell death when co-deleted with another nonessential gene.2 Systematic efforts to determine similar synthetic lethal interactions between pairs of nonessential genes may be especially important in identifying novel targets for cancer therapy. Indeed, it is estimated that each nonessential human gene may have approximately 20 synthetic lethal partner genes.2 Moreover, identification of nonessential genes, which are synthetic lethal in the context of specific cancer drugs, may provide further therapeutic insights. Future efforts utilizing the techniques, tools, and knockout cells created in these studies may provide insight into novel functions of our genome and new therapeutic vulnerabilities to target cancer.
References
Competing Interests
Drs. Taylor and Abdel-Wahab indicated no relevant conflicts of interest.