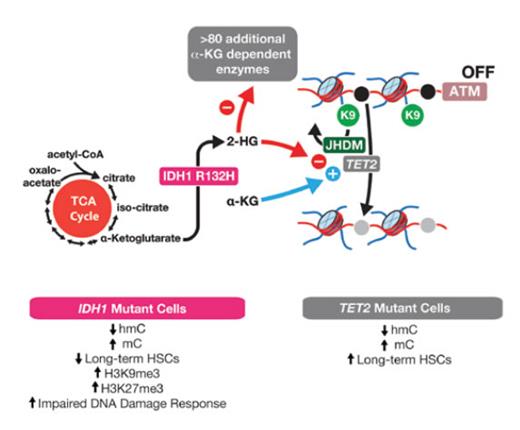
IDH1R132-mutant Cells Have Decreased Ataxia Telangiectasia Mutated (ATM) and Impaired DNA Damage Response, Which Occur Independently From Effects of Mutant Isocitrate Dehydrogenase 1 (IDH) on TET2. IDH1 normally serves in the tricarboxylic acid (TCA) cycle to convert isocitrate to α-ketoglutarate (αKG). Mutations in IDH1 and IDH2 confer a change-of-function of these enzymes causing them to convert αKG to the oncometabolite 2-hydroxyglutarate (2HG). 2HG competitively inhibits αKG-dependent enzymes, which includes the methylcytosine dioxygenase enzyme TET2, the Jumonji C-domain–containing family of histone lysine demethylases (JHDM), and more than 80 additional enzymes. In their article, Dr. Satoshi Inoue and colleagues identify that IDH1R132H-mutant cells have reduced expression of ATM, at least partly due to increased histone H3 lysine 9 (H3K9) trimethylation at the ATM locus. This results in reduced long-term hematopoietic stem cells and impaired DNA damage response in IDH1-mutant cells, which are features not seen in TET2-mutant cells. This stands in contrast to DNA 5-methylcytosine (mC) and 5-hydroxymethylcytosine (hmC), which are similarly altered in TET2- and IDH1-mutant cells.
IDH1R132-mutant Cells Have Decreased Ataxia Telangiectasia Mutated (ATM) and Impaired DNA Damage Response, Which Occur Independently From Effects of Mutant Isocitrate Dehydrogenase 1 (IDH) on TET2. IDH1 normally serves in the tricarboxylic acid (TCA) cycle to convert isocitrate to α-ketoglutarate (αKG). Mutations in IDH1 and IDH2 confer a change-of-function of these enzymes causing them to convert αKG to the oncometabolite 2-hydroxyglutarate (2HG). 2HG competitively inhibits αKG-dependent enzymes, which includes the methylcytosine dioxygenase enzyme TET2, the Jumonji C-domain–containing family of histone lysine demethylases (JHDM), and more than 80 additional enzymes. In their article, Dr. Satoshi Inoue and colleagues identify that IDH1R132H-mutant cells have reduced expression of ATM, at least partly due to increased histone H3 lysine 9 (H3K9) trimethylation at the ATM locus. This results in reduced long-term hematopoietic stem cells and impaired DNA damage response in IDH1-mutant cells, which are features not seen in TET2-mutant cells. This stands in contrast to DNA 5-methylcytosine (mC) and 5-hydroxymethylcytosine (hmC), which are similarly altered in TET2- and IDH1-mutant cells.
One of the most fascinating findings from genomic studies of leukemia from the last 10 years is the discovery of recurrent mutations in isocitrate dehydrogenase 1 (IDH1), IDH2, and ten-eleven translocation 2 (TET2) that have convergent effects on DNA methylation.1 Mutations in IDH1, IDH2, and TET2 are each mutually exclusive of one another and, in total, account for 30 to 60 percent of patients with myelodysplastic syndromes (MDS), acute myeloid leukemia (AML), and other myeloid malignancies. Mutations in IDH1 and IDH2 confer a change-of-function of these enzymes, causing them to convert α-ketoglutarate (αKG) to the oncometabolite 2-hydroxyglutarate (2HG). 2HG competitively inhibits αKG-dependent enzymes, including the methylcytosine dioxygenase enzyme TET2. Since TET2 loss-of-function mutations occur in the same spectrum of hematologic malignancies as IDH1/2 mutations, and because TET2 loss is tightly linked to aberrant self-renewal of hematopoietic stem cells, it has been largely presumed that IDH1/2 mutations promote leukemogenesis by disabling TET2. At the same time, many lines of evidence suggest that IDH1/2 mutations may not be equivalent to TET2 loss. First, there are more than 80 αKG-dependent enzymes beyond TET2. This notably includes the other TET enzymes (TET1 and TET3), the Jumonji C (JmjC) domain–containing family of histone lysine demethylases, and numerous prolyl and lysyl hydroxylases (Figure). Additionally, IDH1, IDH2, and TET2 mutations are each associated with differing clinical outcomes of AML and their own spectrum of coexisting mutations. Therefore, there is a need to dissect potential distinct features of the pathogenesis of IDH- versus TET2-mutant leukemias.
In a recent article, Dr. Satoshi Inoue and colleagues identify impaired DNA damage repair as a unique oncogenic property of IDH1-mutant leukemias, distinct from that of TET2-mutant cells. By performing mass cytometry of hematopoietic cells from Idh1R132H knock-in mice,2 they identified decreased levels of phosphorylated ataxia telangiectasia mutated (ATM) in bone marrow progenitors from Idh1 mutant mice relative to controls. ATM is a kinase that is recruited to double-stranded DNA breaks and plays a role in cell cycle delay during DNA repair. DNA damage accumulated in the progenitor compartment of aged Idh1 mutant mice, but not Tet2 knockout (KO) mice, as noted by increased accumulation of DNA double-stranded breaks, the phosphorylated histone variant H2AX (γ-H2AX), and tumor suppressor p53-binding protein 1 (53BP1). Furthermore, although Idh1 knock-in and Tet2 KO mice both develop a phenotype resembling human MDS, the authors found reduced numbers of long-term hematopoietic stem cells (LT-HSCs) only in the Idh1 knock-in mice. The magnitude of reduction in LT-HSCs also increased with age of the mice, suggesting that decreased ATM impairs the DNA damage response (DDR), leading to the accumulation of unrepaired double-strand breaks over time that potentially trigger apoptosis.
In an attempt to explain the decreased levels of ATM seen in progenitors and LT-HSCs of Idh1 mutant mice, the investigators first looked for DNA hypermethylation at the ATM promoter because IDH1/2 and TET2 mutant cells are marked by DNA hypermethylation. Although there was an increase in DNA methylation at the majority of CpG islands in the genome, there were no significant alterations at the ATM promoter, specifically compared with controls in Idh1 mutant mice. However, chromatin state at the locus of ATM was found to be more compact compared with controls, with an increase in the transcriptionally repressive histone modification histone H3 lysine 9 tri-methylation (H3K9me3). Interestingly, the JmjC-domain–containing family of histone lysine demethylases responsible for removing H3K9 methyl marks is inhibited by 2HG, as noted earlier, thus establishing a possible link between IDH1 and low ATM levels. Furthermore, a pharmacologic inhibitor of H3K9 methyltransferase largely restored ATM mRNA expression.
In Brief
This work provides valuable insights into the biology of mutant IDH1 in a myeloid-specific context with the additive effect of aging, and highlights one of the TET2-independent consequences of 2HG. Given that there are many additional αKG-dependent enzymes beyond TET2 and the JumonjC family of histone lysine demethylases, it is quite likely that there are many additional examples of biological pathways that are specific to IDH1/2-mutant cells. Moreover, distinct clinical associations have been repeatedly observed comparing IDH1R132H-, IDH2R140Q-, and IDH2R172K-mutant patients, possibly related to differences in 2HG produced by each mutation.3 It will therefore be important to determine whether the findings related to impaired DNA damage response in IDH1R132-mutant cells observed here are also seen in IDH2R140Q- and R172K-mutant cells. Finally, inhibitors of mutant IDH1 and IDH2 are now entering phase III testing, with promising results noted in earlier stage trials. However, single-agent therapy does not yet seem to provide durable remissions. If the results from Dr. Inoue and colleagues are reproduced and validated in humans, this finding could offer an opportunity to explore DNA repair inhibitors in combination with IDH inhibitors with the goal of depleting the leukemic stem cell pool to achieve a state of negative minimal residual disease.
References
Competing Interests
Dr. Abdel-Wahab and Dr. Taylor indicated no relevant conflicts of interest.