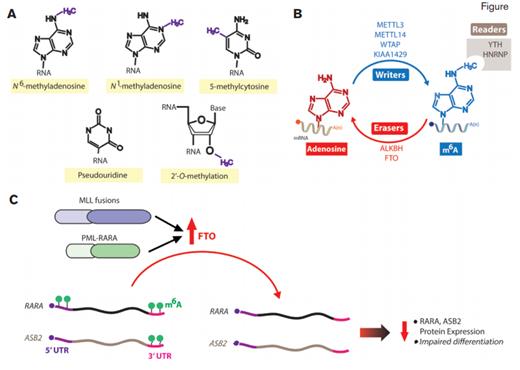
Messenger RNA (mRNA) Modifications, N6-methyladenosine (m6A) Modifiers, and Their Relevance to Acute Myeloid Leukemia (AML). A) Structure of m6A and other mRNA post-transcriptional modifications. B) The writer, reader, and erase proteins of m6A. C) Schema of results from Dr. Li and colleagues who have identified that the m6A eraser FTO is upregulated in MLL- and PML-RARA–rearranged AML and promotes leukemogenesis. FTO removes m6A marks from the 3’ and/or 5’ untranslated regions (UTRs) of select transcripts such as RARA and ASB2, which are important mediators of PML-RARA- and MLL-rearranged leukemogenesis. Removal of these marks was associated with reduced half-life of these transcripts and consequent reduced protein expression and impaired hematopoietic differentiation.
Messenger RNA (mRNA) Modifications, N6-methyladenosine (m6A) Modifiers, and Their Relevance to Acute Myeloid Leukemia (AML). A) Structure of m6A and other mRNA post-transcriptional modifications. B) The writer, reader, and erase proteins of m6A. C) Schema of results from Dr. Li and colleagues who have identified that the m6A eraser FTO is upregulated in MLL- and PML-RARA–rearranged AML and promotes leukemogenesis. FTO removes m6A marks from the 3’ and/or 5’ untranslated regions (UTRs) of select transcripts such as RARA and ASB2, which are important mediators of PML-RARA- and MLL-rearranged leukemogenesis. Removal of these marks was associated with reduced half-life of these transcripts and consequent reduced protein expression and impaired hematopoietic differentiation.
Within the last decade, there has been intense interest in the role of epigenetic alterations, the reversible modifications of DNA nucleotides and amino acids of histone tails, in normal and malignant hematopoiesis. In contrast, there has been very little evaluation of modifications of RNA, which were initially described more than 30 years ago. Analogous to the “readers,” “writers,” and “erasers” of the epigenome, similar proteins that alter and bind RNA modifications have been described (Figure). Collectively RNA modifications are now referred to as the “epitranscriptome” because they can be dynamically placed and removed, just as epigenomic marks on DNA and histones.
The dearth of studies of RNA modifications in cancer to date is explained by the fact that 1) there have been no techniques to map the locations of modifications in RNA until recently, and 2) the proteins that read or alter RNA modifications are rarely mutated in cancer. Moreover, the most common RNA modification, N6-methyladenosine (m6A), constitutes only 0.1 to 0.4 percent of adenosine nucleotides in mammalian cells. Thus, the modest abundance of RNA modifications also has limited efforts to understand the functions of RNA modifications.
Following the description of techniques to map m6A deposition throughout the transcriptome, Dr. Zejuan Li and colleagues now identify that the m6A demethylase FTO (fat mass and obesity-associated protein) promotes acute myeloid leukemia (AML). The authors found that FTO is unique among m6A methyltransferases and demethylases in that its expression is increased in AML over normal hematopoietic cells (Figure). Through a series of functional experiments enforcing expression of wild-type FTO relative to enzymatically dead versions of FTO or RNAi-mediated downregulation of FTO in AML cells, the authors found that overexpression of FTO promotes leukemogenesis of cells bearing fusions of MLL or PML-RARA as well as normal karyotype AML with FLT3-ITD and NPM1 mutations.
Consistent with FTO’s role as an m6A demethylase, FTO overexpression in AML cells was associated with global m6A downregulation. Prior studies have postulated numerous roles for m6A in mRNA metabolism including roles in 5' capping, 3' polyadenylation, splicing, nonsense-mediated decay, and nuclear export of mRNAs. Through correlation of transcriptome-wide maps of m6A deposition (via m6A-seq) with gene expression (via RNA-seq) in cells with and without FTO overexpression, the authors found the somewhat unexpected finding that 80 percent of FTO target mRNAs are negatively regulated by FTO. These data suggest that, at least in the context of AML, FTO-mediated m6A demethylation is a negative regulator of mRNA expression. The authors specifically focused on two m6A methylated genes which appeared to be regulated by FTO, ASB2, and RARA, given prior knowledge of the importance of these genes to MLL- and PML-RARA-rearranged AML. They found that m6A methylation regulates expression of these genes by targeting the 3' untranslated regions (UTRs) of ASB2 and both the 3' and 5' UTRs of RARA (Figure). In fact, modulating the levels of ASB2, RARA, or the enzymes affecting m6A of ASB2 and RARA transcripts regulated differentiation of APL cells to retinoic acid.
In Brief
We are only at the beginning of our understanding of the epitranscriptome in the context of normal and malignant hematopoiesis. Further efforts to map the locations of RNA modifications and decipher the function of readers, writers, and erasers of RNA modifications through deletion and overexpression of these proteins in hematopoietic cells will be critical. Moreover, given the development of therapies targeting so many epigenetic modifiers, similar efforts to develop chemical probes and drugs targeting epitranscriptomic modifiers may be biologically enlightening as well as potentially therapeutically important.
Competing Interests
Dr. Abdel-Wahab indicated no relevant conflicts of interest.