“Alexa, is this a stem cell?”
In the not too distant future, perhaps scientists will be asking their smartphone-run labs this very question regarding a cell in their incubator. Deep learning technologies have recently allowed voice recognition like Amazon’s Alexa or Apple’s Siri to increase their accuracy enough to allow for usable interfaces in real-world situations. This deep learning is not limited to voice, but can be applied to image recognition, game playing, or even driving a car. Just last week, I asked Siri to “find photos of dogs on my phone,” and the software was successfully able to sort through the more than 2,000 images on my phone to find the few that were of our family pet.
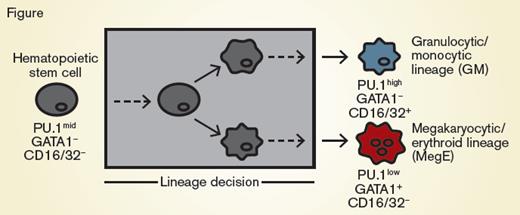
Hematopoietic Stem Cells Differentiating Into GM Cells or MegE-lineage Cells. Hematopoietic stem cells (gray) can differentiate and are annotated as committed toward the granulocytic/monocytic (GM, blue) lineage via detection of CD16/32 or toward the megakaryocytic/erythroid lineage (MegE, red) via detection of GATA1-mCherry expression. These conventional markers appear after the lineage decision of the cell (gray box).
Hematopoietic Stem Cells Differentiating Into GM Cells or MegE-lineage Cells. Hematopoietic stem cells (gray) can differentiate and are annotated as committed toward the granulocytic/monocytic (GM, blue) lineage via detection of CD16/32 or toward the megakaryocytic/erythroid lineage (MegE, red) via detection of GATA1-mCherry expression. These conventional markers appear after the lineage decision of the cell (gray box).
Dr. Felix Buggenthin and colleagues recently applied deep learning processes to hematopoietic stem cell biology. The question they explored is whether deep learning could predict lineage decisions by hematopoietic stem cells. They employed an in vitro culture system such as one previously described,1 where hematopoietic stem cells would differentiate into either granulocytic or monocytic cells, or differentiate into megakaryocytic- or erythroid-lineage cells (Figure). These lineage decisions could be determined downstream by the expression of CD16/32 or the transcription factor PU.1 in the granulocytic/monocytic lineage decision, or by expression of a GATA1-mCherry fusion protein in the megakaryocytic/erythroid lineages. Individual hematopoietic stem cells were cultured, coupled with continuous single-cell imaging. The authors acquired approximately 400 bright-field images per cell throughout the culture, from 5,922 individual cells, resulting in more than 2.4 million images.
The individual hematopoietic stem cells in these cultures either turned on a lineage-restricted expression marker (i.e., were CD16/32 positive or GATA-1 positive), remained lineage marker negative but expressed the marker in a later generation, or never expressed one of the lineage markers. Discarding the images from cells in which no lineage marker was ever expressed, the research team was left with 4,402 analyzed single cells with 1.7 million bright-field images during their culture, with annotation of the lineage choice the cell ultimately made. Applying this dataset to a series of deep neural networks resulted in a computational model that could predict the lineage choice of the cell based on the continuous bright-field images. Remarkably, the model could predict the fate of cells before they expressed any of the lineage markers. In fact, this ability to predict a future lineage choice absent of traditional markers was robust up to three generations before any marker was expressed.
In Brief
The lineage fate choice in this system was hypothesized to be governed by self-reinforcing transcriptional switches, governed by the ratio of PU.1 and GATA-1. However, work published last year1 by this research team challenged that hypothesis and suggested that these transcription factors may simply execute an already decided lineage choice. The ability of the deep learning image analysis to see the future choice of an individual hematopoietic stem cell has immediate applicability to this hematopoietic question. Prospectively identifying a stem cell destined towards a lineage differentiation, up to three generations before that stem cell expresses the transcription factors, can then allow for earlier analysis of transcriptional or epigenetic regulatory elements governing hematopoietic fate. Beyond stem cell biology, we may see similar deep learning image analysis applied to high throughput cancer cell differentiation screens,2 systems exploring cancer resistance mechanisms, or perhaps predicting the best embryos for in vitro fertilization.3 The only question remaining is whether your car will be driving you to your lab before that happens.
References
Competing Interests
Dr. Hoggatt indicated no relevant conflicts of interest.