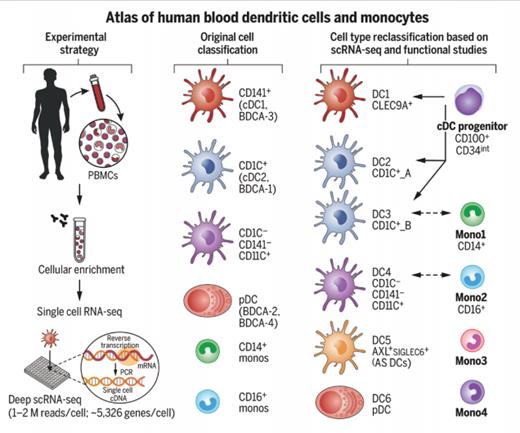
Establishing a Human Blood Monocyte and Dendritic Cell Atlas. Researchers isolated approximately 2,400 cells enriched from the healthy human blood HLA-DR+ lineage- compartment and subjected them to single-cell RNA sequencing. This strategy, together with follow-up profiling and functional and phenotypic characterization, led them to update the original cell classification to include six dendritic cells (DCs), four monocyte subtypes, and one conventional DC progenitor. From Villani AC et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017. doi:10.1126/science.aah4573. Reprinted with permission from AAAS.
Establishing a Human Blood Monocyte and Dendritic Cell Atlas. Researchers isolated approximately 2,400 cells enriched from the healthy human blood HLA-DR+ lineage- compartment and subjected them to single-cell RNA sequencing. This strategy, together with follow-up profiling and functional and phenotypic characterization, led them to update the original cell classification to include six dendritic cells (DCs), four monocyte subtypes, and one conventional DC progenitor. From Villani AC et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017. doi:10.1126/science.aah4573. Reprinted with permission from AAAS.
Taxonomy is a science that struggles to be fashionable. The great evolutionary biologist Steven Jay Gould commented that “Taxonomy is often regarded as the dullest of subjects, fit only for mindless ordering and sometimes denigrated as mere ‘stamp collecting.’” Yet, medicine needs order, and who could deny that the classic chart of “blood cell differentiation,” beloved of scuffed laboratory walls, is imprinted onto the hippocampal map of all hematologists?
Classification systems reflect current technology, and it is no surprise to witness the inexorable dominance of molecular biology. In this remarkable article, Dr. Alexandra-Chloé Villani and colleagues at the Broad Institute deliver a radical revision of the classification of dendritic cells and monocytes.
The breathtaking capabilities of contemporary molecular biology lie at the heart of the analysis. In particular, the work focuses on the use of RNA-Seq, a procedure in which all of the mRNA sequences inside a cell are sequenced such that a complete map of the transcriptional activity can be generated. This technology is the mRNA equivalent of “next generation DNA sequencing” and is rapidly replacing microarray analysis. Perhaps even more remarkable is that this work was done on single cells. This combination of detailed transcriptional assessment and single-cell analysis offers remarkable possibilities for future biological insights. Of the 30,000 genes available within our DNA, around 5,000 are expressed at any time in a single cell, and RNA-Seq normally sequences around 1 million reads such that the technology can discover not only which genes are being expressed but also how many mRNA transcripts are present in the cell.
Dendritic cells (DCs) are relative youngsters within hematopoiesis, characterized by Dr. Ralph Steinman in 1973, and broadly classified into conventional DCs (cDCs), which express CD11c, and CD123+ plasmacytoid DCs (pDCs). cDCs are highly efficient at priming T cell immune responses whereas pDCs are potent producers of interferon-α in response to viral infection. Monocytes are a more familiar feature on our blood smears and have also undergone a binary subdivision through immunophenotyping into classical CD14++ and nonclassical CD14+CD16++ subsets.
In this data-rich but wonderfully accessible article, the authors undertook RNA-Seq on 2,400 single DCs (defined as HLA-DR+ lineage–) and monocytes (CD14+ lineage–) from a single individual. Sequence data were analyzed through a statistical approach called principal component analysis (PCA), which categorized dendritic cells into six major subgroups, while monocytes fell into four subtypes. Surface markers were then used to isolate these subsets, confirm that the cells retained the original RNA profile, and show that the pattern was common in 10 different subjects.
Several novel findings emerge from the reclassification of dendritic cells into six subtypes, termed DC1 to DC6. CD11c+ conventional DCs can be subdivided into those that are CD141+ or CD1C+, or indeed lack both of these molecules. In the new classification, the CD141+ subset becomes DC1 and is renamed CLEC9A+ DC on the basis that CLEC9A is a perfect discriminative marker. The CD1C+ subset is split into two groups, with differential MHC class II or monocytic gene expression (termed DC2 and DC3), while the DC4 group represents the CD1C and CD141 “double negative” group. DC5 is a completely new subset, representing 2 to 3 percent of DCs, and has been termed “AS DC” on the basis of expression of AXL and SIGLEC genes. Finally, DC6 represents the original pDC subset.
Also of note was the finding of a small population of cDC progenitor cells, representing one in 5,000 of the DC population, with a CD100+CD34intermediate phenotype. Morphology plays a role here and shows these cells to possess a high nuclear-to-cytoplasmic ratio with circular or indented nuclei.
The team went on to study monocytes, defined as CD14+lineage–, and delineated four subtypes — two major subsets defined by CD14+ and CD16 expression, and a further two, one with cytotoxic genes and the other with an unknown function.
Several practical lessons are readily apparent from this classification. Functionally, the DC1 through DC5 subsets are capable of stimulating strong T cell responses, whereas DC6 operates primarily for interferon production. The description of a progenitor pool will allow potential expansion of DC cells in vitro with considerable opportunities for genetic manipulation. Additionally, it will prove possible to understand more closely how DC tumors arise during differentiation, and Dr. Villani and colleagues begin this process by showing that the rare condition of blastic plasmacytoid DC neoplasia is most closely related to the pDC (DC6) subset.
In Brief
It should be remembered that this classification will itself ultimately be refined and replaced. It is based solely on transcriptional activity, without regard for features such as phenotype and function, and assesses cells in their resting state, without considering factors such as inflammation. Nevertheless, this report represents a considerable advance in our understanding of these important innate immune subsets. We can now expect this approach to be used for all subsets within the hematologic lineage. That hippocampal map of hematopoiesis is going to get a lot more complicated.
Competing Interests
Dr. Moss indicated no relevant conflicts of interest.