Acute myeloid leukemia (AML) is one of the most challenging malignancies in children and adults due to clinical and biological disease heterogeneity and high relapse rates. Outcomes have improved only modestly with intensive chemotherapy and hematopoietic stem cell transplantation, prompting the ongoing pursuit of immunologically and molecularly targeted therapies. One agent that has been investigated extensively in both children and adults with AML is gemtuzumab ozogamicin (GO), a humanized monoclonal antibody conjugated to calicheamicin that targets delivery of a toxin to CD33-expressing AML cells.
GO initially received accelerated approval by the U.S. Food and Drug Administration (FDA) in May of 2000 as monotherapy for relapsed CD33+ AML in adults, but it was voluntarily withdrawn from the market in 2010 after a subsequent trial failed to confirm clinical benefit, and excessive toxicities were observed. However, there has been renewed interest in GO after more recent studies have demonstrated fewer relapses, better survival, and a favorable safety profile with alternative dosing schedules in subsets of adult patients with de novo AML, particularly those with favorable or normal cytogenetics.1 Notably, on September 1, 2017, the FDA approved GO for the treatment of adults with newly diagnosed CD33+ AML and adults and children at least two years of age with relapsed or refractory CD33+ AML.
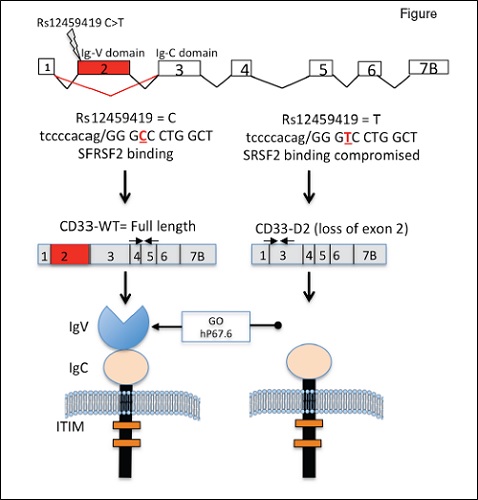
CD33 exon 2 single nucleotide polymorphism (SNP) rs12459419 influences the alternative splicing of CD33. The presence of rs12459419 in exon 2 affects the exonic enhancer binding site for SRSF2, thereby resulting in the loss of exon 2 (shown in red) in the T allele. The loss of exon 2 results in a shorter CD33 isoform lacking the IgV domain, which is recognized by gemtuzumab ozogamicin (GO) and currently used antibodies. Reprinted with permission. © 2017 American Society of Clinical Oncology. All rights reserved. Lamba, JK et al: J Clin Oncol Vol. 35, 2017: 2674-2682.
CD33 exon 2 single nucleotide polymorphism (SNP) rs12459419 influences the alternative splicing of CD33. The presence of rs12459419 in exon 2 affects the exonic enhancer binding site for SRSF2, thereby resulting in the loss of exon 2 (shown in red) in the T allele. The loss of exon 2 results in a shorter CD33 isoform lacking the IgV domain, which is recognized by gemtuzumab ozogamicin (GO) and currently used antibodies. Reprinted with permission. © 2017 American Society of Clinical Oncology. All rights reserved. Lamba, JK et al: J Clin Oncol Vol. 35, 2017: 2674-2682.
The Children’s Oncology Group has conducted three prior trials investigating GO in pediatric patients, the largest of which was the phase III AAML0531 trial.2 Children were randomized to receive conventional chemotherapy versus conventional chemotherapy plus two doses (3 mg/m2) of GO. Improvements in event-free survival and a reduction in relapse risk were observed with the addition of GO. However, responses to GO have been variable, and the potential value of CD33 genotype as a predictive biomarker was demonstrated in this trial. Specifically, CD33 singlenucleotide polymorphism (SNP) rs12459419 C>T (Ala14Val) creates a transcript variant lacking exon 2 (D2-CD33) and eliminating the IgV domain of CD33, the antibody-binding site for GO (Figure). Dr. Jatinder K. Lamba and colleagues evaluated the association between this CD33-slicing SNP and CD33 blast cell surface expression, and outcomes in children treated with and without the addition of GO to conventional chemotherapy.
CD33 coding SNP rs12459419 was genotyped, and CD33 expression levels were determined by measuring blast mean fluorescence intensity by flow cytometry. The authors also assessed CD33 wild-type and alternatively spliced transcript levels from RNA sequencing data obtained from diagnostic patient samples. Among the 816 patients genotyped for rs12459419, the frequencies of the CC, CT, and TT genotypes were 51 percent, 39 percent, and 10 percent, respectively. The authors demonstrated that rs12459419 minor T allele was significantly associated with the expression of an alternatively sliced isoform (D2-CD33) lacking the GO binding site (IgV domain encoded by exon 2). They also showed that the variant T allele was associated with reduced cell surface expression of CD33 assessed by blast mean fluorescence intensity.
Since the rs12459419 variant influences the splice site and results in the expression of a CD33 isoform lacking the GO binding domain, the authors hypothesized that response to GO would be associated with genotype. They correlated the SNP genotype with relapse and response in the GO (n=408) versus non-GO (n=408) treatment arms. Patients with the CC genotype exhibited a significant reduction in relapse rate (26% vs. 49%; p<0.001) and superior disease-free survival (65% vs. 46%; p=0.004) with the addition of GO compared to treatment with conventional chemotherapy alone. While the benefit of GO in patients with the CC genotype was most significant in low-risk patients, modest outcome benefits were also observed in the intermediateand high-risk groups. Moreover, in a multivariable analysis that included CD33 genotype, risk status, and cell surface CD33 expression levels, the rs12459419 CC genotype remained independently associated with response to GO. Conversely, patients with the CT and TT genotypes did not show any benefit from the addition of GO, regardless of risk group.
In Brief
Although GO has re-emerged as a promising AML therapy, responses have been variable. This report highlights the potential value of CD33 genotyping in identifying the patients most likely to benefit from GO. The study findings also have important implications for optimizing the future design of CD33-directed therapies and diagnostic antibodies, the majority of which have historically required the presence of the CD33 IgV domain. The significance of SNP rs12459419 as a response predictor warrants validation in prospective randomized trials in both children and adults.
References
Competing Interests
Dr. Raetz indicated no relevant conflicts of interest.