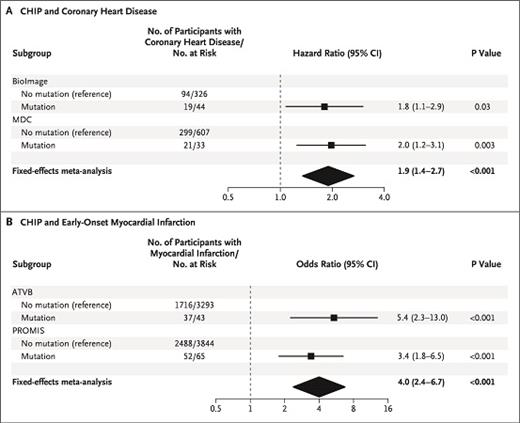
The figure shows the association between clonal hematopoiesis of indeterminate potential (CHIP) and coronary heart disease and early-onset myocardial infarction. Copyright 2017 New England Journal of Medicine. All rights reserved. Jaiswal, S et al: N Engl J Med Vol. 377, 2017: 111-121.
The figure shows the association between clonal hematopoiesis of indeterminate potential (CHIP) and coronary heart disease and early-onset myocardial infarction. Copyright 2017 New England Journal of Medicine. All rights reserved. Jaiswal, S et al: N Engl J Med Vol. 377, 2017: 111-121.
In recent years, high-throughput genome sequencing, or next-generation sequencing (NGS), has become more accessible and affordable, allowing for discoveries of genetic mutations in a variety of clinical settings. This is most commonly used in patients with peripheral blood cytopenias concerning for a myelodysplastic syndrome (MDS). It is not surprising that clonal hematopoiesis — a situation in which hematopoietic stem cells acquire one or more somatic mutations that are detectable by molecular studies — is increasingly being identified. In cases where a definitive diagnosis of a hematologic malignancy such as MDS cannot be made, the significance of clonal hematopoiesis is often unknown.
Peripheral blood cytopenias are commonly found in older individuals and is a typical presentation for MDS. However, many cases of cytopenia remain unexplained after conventional testing and are not attributable to any concomitant disease or medication/toxin effect. The term idiopathic cytopenias of undetermined significance (ICUS) was initially introduced a decade ago to describe cases in which patients have persistent cytopenias, unremarkable peripheral blood and bone marrow morphology, and no MDS-associated cytogenetic abnormalities.1,2 Since then, and with the introduction and increased use of NGS, those with the original definition of ICUS can be further risk stratified based on the presence or absence of a somatic mutation, with those harboring somatic mutations being placed into clonal cytopenia of undetermined significance (CCUS).3,4
Last year, Dr. Luca Malcovati and colleagues demonstrated that in patients with unexplained cytopenias, the detection of somatic mutations can stratify patients as low- or high-risk for developing myeloid malignancies.4 In this study, mutation analysis of 40 selected genes involved in myeloid diseases was performed on peripheral blood of patients with unexplained cytopenias. When a mutation was identified, the patient was moved from ICUS to CCUS, and these patients showed a significantly higher probability of developing a myeloid malignancy. Having a somatic mutation with a variant allele frequency higher than 0.10 or carrying two or more mutations placed patients at a higher risk of developing a myeloid malignancy compared with those having only one mutation or smaller mutant clone. Additionally, not only did the number of mutations and the size of the mutant clone matter, specific mutations such as those in spliceosome genes (SF3B1, SRSF2, and U2AF1) or TET2, ASXL1, or DNMT3A with additional mutations, placed the patient at an even higher risk. These findings can assist in risk stratifying patients into high- or low-risk for developing a myeloid malignancy.
Dr. David Steensma first coined the term “clonal hematopoiesis of indeterminate potential” (CHIP), which is defined as the presence of clonal hematopoiesis in the absence of cytopenias.1 In these individuals, a hematologic malignancy–associated somatic mutation is identified, but they do not have any cytopenias and do not meet the criteria for a hematologic malignancy. It is commonly found in the elderly, it may be found in up to 10 to 20 percent in those older than 70 years, and it is associated with a rate of progression to a hematologic neoplasm of about 0.5 to 1 percent per year.1,4,5
Now, CHIP has expanded into having clinical significance beyond the realm of hematologic neoplasms. In an article published in 2017, Dr. Siddhartha Jaiswal and colleagues discuss the implication of clonal hematopoiesis and the risk for developing atherosclerotic cardiovascular disease.6 The authors used whole-exome sequencing to evaluate for the presence of CHIP in peripheral blood in case-control cohorts. They found that those with CHIP (particularly mutations involving DMNT3A, TET2, ASXL1, and JAK2) were 1.9 times more likely to develop coronary heart disease and four times more likely to develop early-onset myocardial infarction, and hypothesized that it was a causal one. To confirm this hypothesis, the group transplanted atherosclerosis-prone Ldlr knockout mice with bone marrow from TET2 knockout mice and discovered that these mice developed significantly larger atherosclerotic lesions in the aorta compared to control Ldl2 knockout mice who received control bone marrow.
These findings demonstrate that CHIP has clinical impact beyond increased risk for developing hematopoietic neoplasms and may have far-reaching implications in non-hematologic realms. We’ve only begun to scratch the surface in uncovering the clinical consequences of identifying a molecular clone in an otherwise normal individual and in understanding the impact of how it may potentially be used to risk-stratify patients in any number of clinical scenarios.
References
Competing Interests
Dr. George and Dr. Conant indicated no relevant conflicts of interest.