T cell acute lymphoblastic leukemia (T-ALL) is a malignancy of lymphoid origin that accounts for 15 to 25 percent of ALL cases. Survival for patients with both B-cell ALL (B-ALL) and T-ALL has improved substantially throughout the past 30 years, as more than 80 percent of children diagnosed with ALL are cured with modern therapy. Unfortunately, not all patients with T-ALL are cured, and the prognosis for patients with relapsed T-ALL is dismal. More robust genomic characterization of T-ALL blasts has the potential to improve understanding of T-ALL biology, identify novel therapeutic targets, and improve risk stratification. Identifying patients at diagnosis who are likely to relapse could lead to potential modifications in therapy that include intensification of conventional cytotoxic chemotherapy or integration of novel therapies.
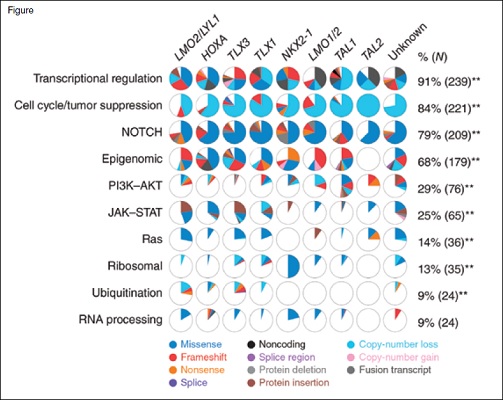
Ten recurrently mutated pathways and the proportion of cases in each T-ALL subgroup shown as pie diagrams. The majority of pathways showed significant associations between prevalence of alteration and T-ALL subgroup; ** p<0.01. Reprinted by permission from Springer Nature: Nature Genetics, The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia, Liu Yu et al, 2017.
Ten recurrently mutated pathways and the proportion of cases in each T-ALL subgroup shown as pie diagrams. The majority of pathways showed significant associations between prevalence of alteration and T-ALL subgroup; ** p<0.01. Reprinted by permission from Springer Nature: Nature Genetics, The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia, Liu Yu et al, 2017.
In 2017, Dr. Yu Liu and colleagues published the largest comprehensive genomic analysis of children and young adults with T-ALL. They performed RNA sequencing, copy number analysis, and whole-exome sequencing on 264 children and young adults with T-ALL who had consecutively enrolled on the Children’s Oncology Group AALL0434 clinical trial (NCT00408005) if both tumor and remission samples were available. They identified 106 putative driver genes, approximately 50 percent of which had not been previously described. They integrated DNA copy number alteration data, sequence mutation, and structural variant/rearrangement, and they identified 10 potentially targetable functional pathways that were recurrently mutated. These included, in order of frequency from most to least common, transcriptional regulation, cell cycle regulation and tumor suppression, Notch1 signaling, epigenetic regulation, PI3K-Akt-mTOR signaling, Jak-Stat signaling, MAPK (RAS) signaling, ribosomal function, ubiquitination, and RNA processing (Figure). They found a strong correlation between the type and frequency of genetic alterations, the developmental stage of T-ALL blasts, and different T-ALL subgroups. For example, alternations in PI3K/Akt/mTOR signaling were prevalent in the TAL1 subgroup, whereas alterations in Jak-Stat and MAPK signaling were prevalent in the LMO2/LYL1 and HOXA subgroups. Finally, they established that multiclonal and subclonal mutations were common.
While the authors made many seminal observations with this groundbreaking study, there were a few limitations to the analysis. First, only a small percentage of samples included whole-genome sequencing. Numerous recent studies have demonstrated the importance of genomic alterations in noncoding regions in T-ALL pathogenesis.1,2 Second, only 7.5 percent of the patients included in the cohort relapsed. Thus, the study was not powered to correlate genetic lesions with outcome. A cornerstone to the improvement in ALL survival has been patient risk stratification based on sentinel genetic alterations and measurement of minimal residual disease (MRD). While this strategy has been effective for B-ALL, it has been less effective for patients with T-ALL because no known genetic alterations have been demonstrated to be independently prognostic of MRD. While MRD is successful in identifying most T-ALL patients who have a worse outcome, most relapses occur in patients classified as lower risk by MRD. Comprehensive genomic analysis of a larger population of relapsed patients has the potential to identify genetic alterations or groups of alterations that may predict outcome. Lastly, one of the criteria for inclusion in the analysis was availability of a remission sample for germline DNA. This excluded some patients who were chemorefractory (i.e., had induction failures).
In Brief
In summary, Dr. Liu and colleagues performed a robust, comprehensive genomic analysis of patients with T-ALL. They made many important observations that are directly translatable and will improve understanding of T-ALL biology. Hopefully, these observations will lead to the development of new therapies that may improve survival for children and young adults with T-ALL.
References
Competing Interests
Dr. Teachey indicated no relevant conflicts of interest.