The ability to diagnose disease accurately and to determine the most effective therapy based on key determinants of outcome in a holistic manner is the holy grail of precision medicine. For diffuse large B-cell lymphoma (DLBCL), the most common and aggressive form of non-Hodgkin lymphoma, accurate diagnosis is usually straightforward; however, our allocation of therapy remains suboptimal for the approximately 40 percent of patients not cured by standard R-CHOP (rituximab + cyclophosphamide, doxorubicin, vincristine, and prednisone) chemoimmunotherapy. Better discrimination of the determinants of outcome beyond the clinically based international prognostic index (IPI) score and more effective therapies for poor-outcome subgroups are needed.
Risk stratification of patients with DLBCL has become more sophisticated in recent years. When considered in addition to the robust IPI, biological classifiers such as cell-of-origin and MYC/BCL2/BCL6 translocation status have added some value. Despite this, distinct treatment paths for biological subgroups are not routinely considered.
Furthermore, the advent and increasing use of massively parallel sequencing (MPS) has implicated somatic alteration in a large number of genes in the pathophysiology of DLBCL. Until recently, however, genomic landscaping studies have been either insufficiently powered or have lacked the clinical annotation required to comprehensively define distinctive clinicopathological subgroups within the heterogeneity observed in DLBCL.
The study by Dr. Roland Schmitz and colleagues sought to explore the genomic landscape of DLBCL and to map specific combinations of genetic changes onto cell-of-origin categories defined by gene-expression profiling. A total of 574 prospectively collected and frozen DLBCL samples underwent comprehensive, multiplatform genomic analysis (whole-exome, targeted amplicon and transcriptome sequencing). The cohort was composed of the following cell-of-origin subgroups: activated B-cell–like (ABC; 51.4%), germinal-center B-cell–like (GCB; 28.6%), and unclassified (20.0%).
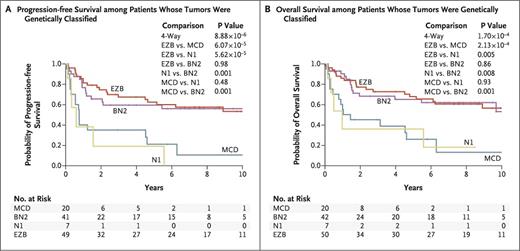
Numerous genes altered either by somatic mutation and/or copy number variation (CNV) were significantly differentially distributed among the three cell-of-origin categories. This observation formed the basis of a new, validated genomic classifier consisting of four novel subgroups of DLBCL that accounted for 44.8 percent of cases in the cohort: MCD, based on MYD88 L265P and CD79B co-mutation; BN2, based on BCL6 fusion and NOTCH2 mutation; N1, based on NOTCH1 mutation; and EZB, based on EZH2 mutation and BCL2 translocation.
Three of these genomic subgroups were highly correlated with cell-of-origin gene-expression categories: MCD and N1 with the ABC category, and EZB with the GCB category. By examining the pattern of recurrent gene aberrations as well as differential gene-expression data, Dr. Schmitz and colleagues were able to identify the cellular pathways most engaged by each genomic subgroup beyond the defining lesions described above, including in many of those unclassifiable by cell of origin.
In the MCD subgroup, dysregulation of plasmacytic differentiation, tumor suppressor gene loss, immune editing, NK-cell anergy, and an IRF4-mediated ABC gene-expression profile were prominent features. On the opposite end of the cell-of-origin spectrum, the EZB subgroup was characterized by REL amplification, tumor suppressor loss, disruption of germinal center homing, immune editing, enhanced JAK-STAT, and PI3 kinase signaling, as well as a BCL6/TCF3–mediated GCB gene-expression profile.
The other two genomic subgroups are characterized by NOTCH gene mutations and expression signatures. However, NOTCH1 mutations (which characterize the N1 subgroup) and mutations of NOTCH2 (which typify the BN2 subgroup) and the related gene SPEN were mutually exclusive, suggesting distinct pathophysiologies. The BN2 subgroup was enriched for NOTCH and NF-κB pathway aberrations while the N1 subgroup contained aberrations affecting B-cell differentiation, which may account for the plasma-cell and quiescent-cell gene-expression signatures observed in this subgroup.
From New England Journal of Medicine, Schmitz R et al, Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma, Vol 378, 1396-1407. Copyright 2018. Reprinted with permission from Massachusetts Medical Society.
From New England Journal of Medicine, Schmitz R et al, Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma, Vol 378, 1396-1407. Copyright 2018. Reprinted with permission from Massachusetts Medical Society.
Clinical outcomes data were available in 240 cases of untreated DLBCL receiving R-CHOP or similar chemoimmunotherapy. Approximately half of these patients could be assigned a genomic subgroup. The ability of the genomic classifier to predict for patient outcome was striking (Figure), with the BN2 and EZB subgroups enjoying significantly superior survival rates (65% and 68% 5-year overall survival [OS], respectively) when compared with the MND and N1 subgroups (26% and 36% 5-year OS, respectively).
Of note, the genomic classifier significantly improved prognostication from diagnosis when added to either the IPI or cell-of-origin classifier.
In Brief
This ground-breaking work by Dr. Schmitz and colleagues has significantly enhanced our understanding of DLBCL pathophysiology and refined our ability to risk stratify patients prior to first-line chemo-immunotherapy. The validation of these results in larger cohorts is necessary, and there is clear need for ongoing work in this area, as more than half of DLBCL cases could not be genomically classified. However, a comprehensive clinicogenomic classifier that considers the IPI, cell-of-origin, and genomic subgrouping concurrently is likely to provide the best possible risk stratification of DLBCL. Indeed, another recently performed landmark genomic analysis study of DLBCL by Dr. Anupama Reddy and colleagues has recently resulted in a web-based tooltool that attempts this very thing.1
The significance of defining distinct genomic subgroups reaches well beyond the prediction of response to first-line chemoimmunotherapy. The significant risk of relapse in DLBCL, especially in patients with high IPI scores, necessitates the pursuit of rationally selected new therapies. For example, Bruton tyrosine kinase inhibitors such as ibrutinib are reported to have significant efficacy in ABC DLBCLs with both MYD88 L265P and CD79B mutations. The association of these aberrations with the MCD subgroup, together with the higher frequency of aberrations involving the B-cell receptor–dependent NF-κB pathway in the MCD and BN2 versus N1 and EZB subgroups, implies potential differential response to BTK inhibition across the genomic subgroups.
The consideration of genomic subgrouping in the design of future trials of targeted agents in DLBCL may allow for the rational selection of patients most likely to derive benefit. While trials of “all-comers” may prove to be negative, those enriched for patients with the appropriate biology may potentially pave the way for a therapeutic paradigm shift in this disease. However, caution is warranted as not every targetable lesion will lead to disease response when hit, particularly in a complex disease such as DLBCL that can use multiple oncogenic pathways of drug escape. This was well illustrated in the aforementioned study by Dr. Reddy and colleagues, where CRISPR-based knockout of certain targetable lesions such as NOTCH2 did not have an effect on DLBCL cell proliferation in model systems.
In summary, the study by Dr. Schmitz and colleagues represents a major advancement of our biological and clinical understanding of DLBCL and represents a meaningful step toward the systematic evaluation of the role of precision medicine in this disease. However, more work is required to enable the comprehensive clinicogenomic classification of a greater proportion of cases and to lower the practical barriers to accessing sophisticated, real-time genomic testing outside highly specialized academic settings.
References
Author notes
Editor's Note: The articles by Drs. Jacobson and Roberts in this issue discuss two related publications in the exciting area of the genetic classification of diffuse large B-cell lymphoma, albeit from different angles. For coverage of the article by Chapuy et al, as covered by Dr. Jacobson, read her DiffusionDiffusion.
Competing Interests
Dr. Yannakou and Dr. Roberts indicated no relevant conflicts of interest.