The explosion of next-generation gene sequencing (NGS) in the last five years has provided major new insights into the biology of acute myeloid leukemia (AML) while posing new dilemmas for hematologists in both the laboratory and the clinic. Two discoveries have been particularly confronting. First, healthy older people with normal blood counts often can harbor myeloid clones bearing mutations in AML-driver genes — so-called clonal hematopoiesis of indeterminate potential (CHIP) or age-related clonal haematopoiesis, as reviewed by Drs. Joanna Conant and Tracy George in the 2018 Year’s Best issue of The Hematologist.1 Second, many AML patients in first remission have persisting clonal hematopoiesis–bearing mutations in AML-driver genes, as discussed by Dr. Annette Kim in a July/August 2018 Diffusion article.2 These discoveries pose the obvious question of what the detection of mutant myeloid clones means for healthy people and whether all clones have similar significance.
This year, major advances have been made in deciphering these questions in the context of healthy individuals with no history of blood disorders, and two articles are equally worthy of being listed as the year’s best. Dr. Sagi Abelson and colleagues3 and Dr. Pinkal Desai and colleagues4 both tackled the challenge of defining the risk of developing AML when mutant myeloid clones are detectable in healthy people. Dr. Abelson and colleagues identified 95 individuals with blood samples suitable for sequencing an average of 6.3 years before AML diagnosis (pre-AML group) and compared their mutational profile with that of 414 unselected age- and sex-matched controls. Dr. Desai and colleagues performed deep sequencing of serially collected peripheral blood samples obtained from 212 women a median of 9.6 years before their diagnosis of AML, along with 212 age-matched controls. Reassuringly, the results of both studies are generally consistent and informative.
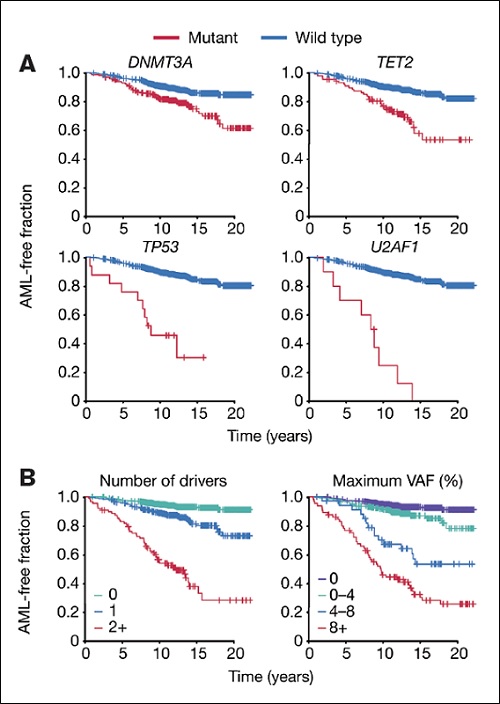
Kaplan-Meier curves of acute myeloid leukemia (AML)-free survival, defined as the time between sample collection and AML diagnosis, death or last follow-up. Survival curves are stratified according to mutation status for selected genes (A) or number of driver mutations per individual and largest clone detected (B). Adapted/Translated by permission from Springer Nature: Abelson S. et al. Prediction of acute myeloid leukaemia risk in healthy individuals. 2018;559:400-404.
Kaplan-Meier curves of acute myeloid leukemia (AML)-free survival, defined as the time between sample collection and AML diagnosis, death or last follow-up. Survival curves are stratified according to mutation status for selected genes (A) or number of driver mutations per individual and largest clone detected (B). Adapted/Translated by permission from Springer Nature: Abelson S. et al. Prediction of acute myeloid leukaemia risk in healthy individuals. 2018;559:400-404.
Compared to controls, patients destined to develop AML were more likely to have a detectable mutation in an AML-driver gene (odds ratio, 4.86 in Dr. Desai and colleague’s study) extending previous data that indicated a higher risk of developing myeloid malignancy, but not AML specifically, for people with CHIP. However, CHIP was common in controls, especially in those older than 65 years (>30% in both studies), and AML was rare, meaning that most people with CHIP are not destined to develop AML. To address this, both studies refined their analyses, focussing on comparisons between people with pre-AML and controls who had CHIP. They identified that AML risk increased as the number of mutations increased, as clone size (measured as variant allele frequency [VAF]) increased and if the driver mutations were in specific genes, particularly TP53, splicing factors (e.g., U2AF1, SRSF2), DNMT3a, TET2, and, in Dr. Desai’s study only, IDH1, IDH2, and SF3B1 (Figure). TP53 mutations conferred the highest risk when genes were considered in isolation. The most commonly aberrant CHIP-defining genes, DNMT3a and TET2, were associated with heightened risk of progression to AML as single abnormalities, but most healthy people with these mutations as single aberrations did not progress to AML. The risk was much greater when more than one variant was detected and when DNMT3a mutations co-occurred with a splicing gene mutation. These two studies now allow identification of people with CHIP who are at highest risk of subsequently developing AML. Importantly, they teach us that when assessing a person with CHIP, the details contained within the molecular pathology report are crucial.
Also crucial is clinical context. Patients with AML in first complete remission (CR) and their physicians are increasingly turning to minimal residual disease (MRD) assessment to assist in decision-making about post-remission care, yet when the same NGS techniques are applied, many patients have detectable mutant myeloid clones when in CR. Two articles in the past year have illuminated this issue.5,6 In both, paired bone marrow samples (before and after remission induction) were analyzed using targeted NGS panels and MRD status assigned based on the presence or absence of detected somatic mutations in the post-treatment CR samples. Results were correlated with clinical outcomes such as relapse rate and overall survival. The findings across both studies were similar.
Mutations identified at diagnosis were detected in remission in approximately 50 percent of patients in each study. While mutations in particular genes (e.g., NPM1, FLT3, CEBPA) were very likely to be cleared in CR, others were not. Mutations in three genes (DNMT3a, TET2 and ASXL1; so called “DTA” mutations ) commonly mutated in both AML and CHIP normally persisted in CR. In Dr. Mojca Jongen-Lavrencic and colleagues’ study, the persistence rates for DTA mutations were 78.7 percent for DNMT3a, 54.2 percent for TET2, and 51.6 percent for ASXL1. Persistence of DTA mutations was not associated with an increased risk of relapse over three years of follow up. In sharp contrast, persistence of non-DTA mutations was highly correlated with risk of relapse and death, and as with CHIP in healthy people, the size of the mutant clone was important. Therefore in the context of AML in first CR, the clinical importance of detecting a persisting mutant myeloid clone pivots on which gene or genes are mutated. As in the case of CHIP in healthy people, the presence of a clone with a single mutation in DNMT3a or TET2 seemed compatible with a favorable outcome in the first three years of follow up.
Collectively, the findings of these articles provide valuable clues as to how to assess what a mutant myeloid clone means for a specific patient. The gene(s) affected, the number of mutations, the size of the clone, and clinical context, all need to be considered together.
References
Competing Interests
Dr. Roberts indicated no relevant conflicts of interest.