Inhibitors of the two isoforms of isocitrate dehydrogenase (IDH), the cytosolic form IDH1 and the mitochondrial form IDH2, were first-in-class compounds with novel allosteric mechanism of action.1,2 The wild-type (WT) versions of these enzymes play a critical role in the citric acid cycle, in the conversion of isocitrate to α-keto-glutarate (αKG). However, mutations in the genes encoding these enzymes have been identified in myeloid neoplasms, gliomas, chondrosarcomas, and intrahepatic cholangiocarcinomas. These hotspot missense mutations, all affecting arginines (IDH1 p.R132 and its cognate IDH2 p.R172 as well as IDH2 p.R140), are located in the enzymatic active site and are critical residues that interact with the substrate, resulting in the formation not of αKG, but of the oncometabolite (R)-2-hydroxygluarate (2HG) that deregulates the epigenetic landscape of the cancer cell. Despite the presence of a catalytic site, the IDH1 and IDH2 inhibitors ivosidenib and enasidenib- respectively, bind to the protein-protein interface formed by homodimerization required for enzymatic function.3
In some patients, these compounds have produced dramatic and durable responses.1,2 However, it should come as no surprise that, as with any chronically administered oncologic agent, patients have developed resistance to the drugs over time. Analogous to the various mutations that have been identified in patients on tyrosine kinase inhibitors such as imatinib that prevent binding of the drug to its target at/near the ATP binding pocket, Dr. Andrew Intlekofer and colleagues at Memorial Sloan Kettering Cancer Center have identified mutations at the binding site of the IDH inhibitors. However, since these are allosteric compounds, the resistance mutations are correspondingly at the site of protein-protein interactions. Importantly, these mutations would therefore be missed by the many allele-specific assays and hotspot-directed targeted next-generation sequencing panels.
Dr. Intlekofer and colleagues identified two sentinel patients who developed resistant acute myeloid leukemia (AML) while on enasidenib and were found to have IDH2 p.Q316E and p.I319M mutations, respectively. In contrast to the initial pathogenic mutation at p.R140Q in exon 4, these two secondary mutations are located in exon 7, which encodes the dimerization interface. These mutations were not identified prior to treatment with enasidenib (at the analytical sensitivity of digital droplet polymerase chain reaction [ddPCR]), but only after six or nine months of enasidenib. Individual patient IDH2 alleles were amplified by long-range PCR and subcloned. Sequencing of resultant colonies definitively demonstrated that the p.R140Q and p.Q316E or p.I319M alleles were identified in trans. Engineered double mutants (p.R140Q/p.Q316E or p.R140Q/p.I319M) and the single mutant (p.R140Q/WT) in cell lines, mice, and in in vitro experiments with purified enzyme all showed that while p.R140Q/WT was sensitive to enasidenib, the double mutants demonstrated resistance to IDH2 inhibition, as measured by 2HG production, NADPH production, and/or colony formation ability.
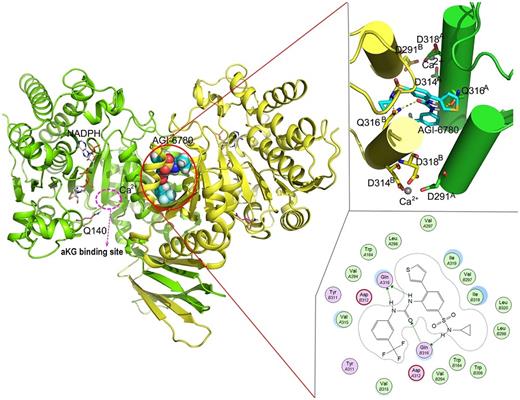
Structure of an IDH2 inhibitor (AGI-6780, structurally related to enasidenib and ivosidenib) allosterically bound at the homodimerization interface with detailed interaction diagrams to the right. Note the location of the Ile 319 and Gln 316 in the bottom right with critical hydrogen bonds to Gln 316. (Reprinted with permission from Chen J, Yang J, Sun X, et al. Allosteric inhibitor remotely modulates the conformation of the orthestric pockets in mutant IDH2/R140Q. Sci Rep. 2017 Nov 28;7:16458. http://creativecommons.org/licenses/by/4.0/)
Structure of an IDH2 inhibitor (AGI-6780, structurally related to enasidenib and ivosidenib) allosterically bound at the homodimerization interface with detailed interaction diagrams to the right. Note the location of the Ile 319 and Gln 316 in the bottom right with critical hydrogen bonds to Gln 316. (Reprinted with permission from Chen J, Yang J, Sun X, et al. Allosteric inhibitor remotely modulates the conformation of the orthestric pockets in mutant IDH2/R140Q. Sci Rep. 2017 Nov 28;7:16458. http://creativecommons.org/licenses/by/4.0/)
Interestingly, structural modeling of the homodimer interface with enasidenib demonstrate that, while the interface is symmetric, since enasidenib is not, the same residues on either side of the drug will affect binding differentially. While the p.Q316E mutation results in electrostatic disruption of a critical hydrogen bond, the p.I319M mutation sterically inhibits the binding of compound (Figure).3 This raised the interesting possibility that double mutations in cis might also result in secondary resistance. The authors therefore searched patients with either primary or secondary resistance to either ivosidenib or enasidenib for cis double mutations and were able to identify a single patient with an IDH1 p.R132C/p.S280F cis double mutation (the S280 residue is paralogous to the I319 residue in IDH2), confirming their hypothesis. It is unclear whether these are the only residues affected, or whether, as in the case of TKI resistance, many other potential secondary resistance mutations will be found.
The same group, with primary author Dr. James Harding, went on to identify an orthogonal mechanism for resistance to IDH inhibition —isoform switching. This group identified four patients, representing a mixture of myeloid and solid tumor neoplasms, in whom the clonal evolutionary pressure of inhibition of one mutated IDH isoform resulted in outgrowth of the neomorphic version of the other IDH isoform. In these patients, relapse/progression was noted at approximately a year or more after treatment, but in some cases, the new mutation was present at low levels before treatment with the initial inhibitor, while in others it emerged soon after the start of therapy. Significantly, the isoform switching was found to be bidirectional (new IDH1 mutation in enasidenib or new IDH2 mutation on ivosidenib). This escape mechanism was also documented in two of 12 cases (17%) of resistance to enasidenib therapy with emergence of IDH1 mutations.4 From the available literature, it is yet unclear if these different isoform mutations represent separate clones or a subclonal event.
In Brief
In summary, Dr. Intlekofer and colleagues identified two orthogonal mechanisms for evasion of IDH inhibition, either by the development of secondary resistance mutations at the homodimerization allosteric binding site of the drugs in cis or in trans, or through bidirectional isoform switching. The former study illustrates an important novel mechanism for the escape of the neoplastic clones that have broad implications in the realm of hematologic neoplasms as well as solid tumors, as there are many potential homomultimeric targets (e.g., tyrosine kinases) in cancer biology. The study by Dr. Harding and colleagues raises the possibility of dual IDH inhibition at the outset of therapy to prevent clonal evolution and evasion. Lastly, there are important pathology ramifications of these studies. They highlight the importance of whole coding sequence coverage of genes of targeted therapies, rather than hotspot single gene assays or multigene panels, as well as the importance of coverage of relevant biologic isoforms as well.
References
Competing Interests
Dr. Kim indicated no relevant conflicts of interest.