Recurrent genetic abnormalities in acute myeloid leukemia (AML) confer distinct clinicopathologic characteristics and are the foundation of modern AML subclassification systems. Among the most common recurrent genetic driver mutations in AML are activating internal tandem duplications (ITD) within the juxtamembrane domain of the type 3 receptor tyrosine kinase FLT3 (FLT3-ITD) and exon 12 frameshift mutations in nucleophosmin 1 (NPM1).1 FLT3-ITDs are present in 25 percent of all AML cases and are also found in a subset of acute lymphocytic leukemia (ALL), while NPM1 mutations are more restricted to AML and occur in 35 percent of cases. NPM1-mutated AML is generally associated with a favorable prognosis, while the presence of a FLT3-ITD generally confers a poor prognosis, especially at a high (>0.5) FLT3-ITD allele fraction.
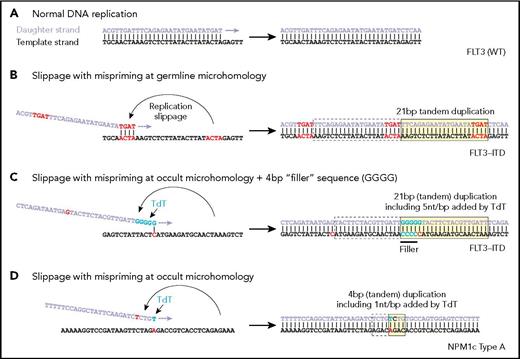
Examples of replication showing (A) normal DNA replication, (B) microhomology resulting in mispriming and formation of an internal tandem duplication (ITD), (C) nontemplated addition of nucleotides by TdT and the use of the terminal nucleotide to create an occult single base mispriming event to create an ITD with filler sequence, and (D) a similar occult mispriming event leading to an NPM1 four base pair insertion. (From Vassiliou GS. The curious incident of TdT-mediated mutations in AML. From Vassiliou GS. The curious incident of TdT-mediated mutations in AML. Blood 2019;134:2229-2231.
Examples of replication showing (A) normal DNA replication, (B) microhomology resulting in mispriming and formation of an internal tandem duplication (ITD), (C) nontemplated addition of nucleotides by TdT and the use of the terminal nucleotide to create an occult single base mispriming event to create an ITD with filler sequence, and (D) a similar occult mispriming event leading to an NPM1 four base pair insertion. (From Vassiliou GS. The curious incident of TdT-mediated mutations in AML. From Vassiliou GS. The curious incident of TdT-mediated mutations in AML. Blood 2019;134:2229-2231.
Although the molecular events underlying the formation of NPM1 mutations and FLT3-ITDs are poorly understood, DNA polymerase slippage near repetitive (microhomology) sequences has long been speculated as a culprit for FLT3-ITDs.2 A study published recently by Dr. Julian Borrow and colleagues analyzed 300 FLT3-ITDs from 271 patients for sequence-level evidence of polymerase slippage. The researchers hypothesized that if this mechanism were responsible for ITD formation, the sequences immediately flanking the 3′ and 5′ ends of the duplication would include short (1-6 base pairs), homologous sequences (microhomology), which is indeed seen in a minority of cases (Figure 1A compared to 1B).3 Unexpectedly, microhomology was not observed in a majority (62%) of ITDs. In fact, 38 percent of all ITDs contained an additional sequence of unknown origin (nontemplated “filler”) with the remaining 24 percent containing neither overt microhomology nor filler.
To explain the frequent presence of filler in FLT3-ITDs, the authors use a series of mathematical deductions and inferences to suggest an unusual mechanism: activity in the myeloid cells of terminal deoxynucleotidyl transferase (TdT), the DNA polymerase responsible for untemplated additions of nucleotides to V, D, and J exons during immunoglobulin and T-cell receptor rearrangement. Although canonically thought of as restricted in expression to lymphoid cells, TdT is also known to be expressed in a subset of AML at diagnosis and in myeloid progenitors. Supporting the involvement of TdT in FLT3-ITD formation, filler sequences were found to be enriched for G/C content at a level consistent with TdT’s known bias with filler sequences of a length consistent with TdT activity. Dipurines and dipyrimidines were observed at a higher than expected frequency in filler sequences, as would be expected during nontemplated polymerization by TdT. Finally, filler sequence correlated with frequency of TdT expression across AML subtypes and was significantly overrepresented in FLT3-ITDs derived from ALL compared to AML.
The authors went on to show how TdT activity could also provide a basis for generation of ITDs in the absence of germline microhomology. In short, TdT-added nucleotide runs that, by chance, have homology with nearby 5′ sequence (occult microhomology, Figure, part C) could allow for polymerase repositioning and subsequent duplication. These runs would not be apparent as filler sequence, but they should display the level of G/C bias associated with TdT activity. In a companion study, Dr. Borrow and colleagues extended their sequence analysis approach to 2,430 previously published NPM1 mutations in AML (114 unique mutations), showing that of 114 unique mutations, more than 90 percent of NPM1 mutations, contained filler sequences that were consistent with TdT activity in terms of G/C and dinucleotide bias as well as length distribution (Figure, part D).
In Brief
In summary, Dr. Borrow and colleagues demonstrated sequence-level evidence that TdT plays a causal role in the generation of FLT3-ITD and NPM1 mutations in AML, perhaps explaining their common co-occurrence. However, the clinical context and prognostic value of filler and microhomology status in FLT3-ITDs and NPM1 mutations remain to be determined. Most AMLs lack expression of TdT by immunohistochemical stains or flow cytometry. The authors have suggested that TdT must be expressed early in the leukemic stem cell, while the block in maturation is expressed slightly later, after down-regulation of the enzyme. One would assume that similarly, NPM1 mutations should also occur with greater frequency in cases with earlier block in maturation, although experientially, NPM1 is more common in AMLs with some degree of differentiation (especially monocytic). The prognostic implications are even more intriguing. While allele fraction is perhaps the most widely recognized stratification for survival within FLT3-ITD+ AML,1 a recent study demonstrated that FLT3-ITDs containing filler sequences are associated with poor response to both cytotoxic and tyrosine kinase inhibitor therapy, suggesting a potential role for these structural characteristics as predictive biomarkers.4 Studies on the differential behavior of NPM1 insertion sequences are more aspirational at this time. In combination, these studies suggest that the “gold standard” capillary electrophoresis sizing assays for FLT3 and NPM1 may be insufficient in the future to appropriately stratify these patients, and that typing of the actual insertion sequence will be critical. This study is the first to implicate TdT in carcinogenesis and opens the door to future searches of the cancer genome for the polymerase’s improvisational yet productive mark.
References
Competing Interests
Dr. LaMacchia and Dr. Kim indicated no relevant conflicts of interest.