Editor’s Note
The first mention of clonal hematopoiesis (CH) of indeterminant potential as a named entity appeared in PubMed in 2015. Since then, this condition has been the subject of numerous, critically important investigations. In this Mini Review, Drs. O’Sullivan and Mead highlight two recent publications that expand what we know about the relationship between CH and the development of outright hematologic malignancies.
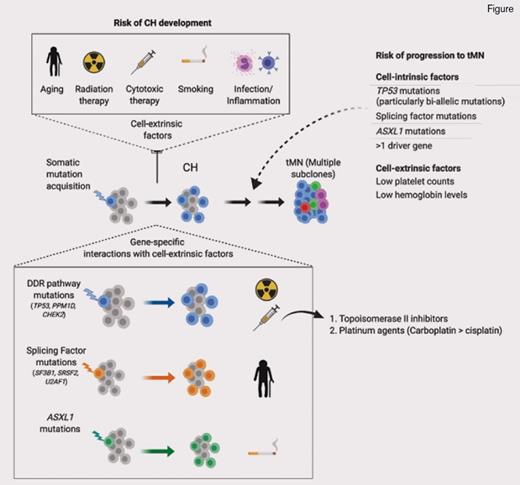
CH is a well-recognized entity where a somatic mutation is acquired by a single hematopoietic stem cell, confers a fitness advantage, and leads to clonal expansion. Eventually, the hematopoietic stem cell–derived clone expands such that it contributes to a considerable proportion of mature blood cell production in apparently healthy individuals. CH is more prevalent with increasing age and carries an increased risk of later evolution to a myeloid neoplasm (MN).1,2 Mutations most often detected in CH mirror those seen in overt MNs; these include epigenetic modifying genes (DNMT3A, TET2, ASXL1, IDH1/2), splicing factors (SF3B1, SRSF2, U2AF1), and DNA damage response (DDR) pathways (TP53, CHEK2, and PPM1D). MN-associated signaling pathway mutations are less frequent in CH, except for the JAK2 V617F mutation. Numerous studies have helped to refine the risks of transformation in CH,1,2 and a crucial role for mutations affecting DDR pathways is emerging. Several important studies have described that TP53 or PPM1D mutation–associated CH is particularly common after previous cancer treatment.1-3 Furthermore, MNs arising after chemotherapy or radiotherapy (therapy-MNs [tMNs]) are also enriched for TP534 and PPM1D5 mutations, which are often present prior to cancer treatment.6
TP53, a tumor suppressor gene, is the most commonly mutated gene across all cancers, and in MNs is associated with very poor outcomes and resistance to standard treatments.7 When present in patients with tMNs, TP53 mutation is associated with a similarly dismal prognosis.8 Therefore, new approaches for early detection and prevention of TP53-associated tMN are badly needed. However, not all patients with TP53 mutation develop tMNs. So how do we identify patients who might be at particular risk of tMNs through cell-intrinsic or cell-extrinsic factors? Regarding cell-intrinsic factors, a wide spectrum of different TP53 mutations can occur as mono- or biallelic mutations. Whether such TP53 mutant allelic imbalance has important clinical implications remains unclear, as monoallelic TP53 mutation can mimic TP53 loss of function by exerting a dominant negative effect.9
Hoping to shed light on this question, Dr. Elsa Bernard and colleagues10 recently reported an analysis of TP53 mutations in myelodysplasia (MDS) patients (n=3,324), inclusive of a subgroup with therapy-related MDS (n=229). They studied the TP53 allelic state using a combination of conventional G-banding analyses and a next-generation sequencing (NGS) panel covering TP53 and genomewide copy-number probes. One-third of TP53 mutant cases of MDS were monoallelic and two-thirds had multiple hits, consistent with biallelic mutation. Variant allele frequency (VAF) measurements for most cases correlated with monoallelic or biallelic states. However, some patients with VAF below 50 percent (n=19/378 patients) had copy neutral loss of heterozygosity at the TP53 locus and would have been miscategorized as monoallelic if based on VAF alone. Therefore, VAF should not be used as the sole method of assigning allelic state.
Biallelic TP53 mutations were associated with poorer survival and an increased risk of acute myeloid leukemia (AML) transformation in contrast to persons with monoallelic TP53 mutation where the outcome was comparable to that of TP53 wild-type patients. Serial sample analysis in patients with AML transformation detected a higher proportion of biallelic TP53 alteration at the time of transformation, underlining the key role for loss of wild-type TP53 as the event that drives disease progression. Previous studies have also reported better survival in MDS patients with TP53 mutation with VAF below 20 percent (monoallelic for the majority).11 However, analyses in AML found TP53 mutations to be associated with adverse prognosis irrespective of VAF.12
Patients with tMNs in this study10 had a higher frequency of multiple mutations of TP53 as compared with de novo cases (84% vs. 65%; OR, 2.8; p=0.002). The same observations for clinical outcomes were made in this tMN subgroup; those with biallelic mutations had poorer outcomes compared to those with monoallelic TP53 mutations who had a lower risk of death, though this did not achieve statistical significance.
The relationship between CH associated with DDR mutations, chemotherapy treatment, and risk of tMN was studied in greater depth by Dr. Kelly Bolton and colleagues.13 Their group set out to determine novel approaches that might be used for early intervention and targeted prevention for patients with cancer at high risk of developing tMN. They analyzed targeted, deep coverage NGS data (MSK-IMPACT) to detect CH in a very large cohort of patients with cancer (n=24,439). Strikingly, the authors reported a prevalence of CH mutations in almost one-third of patients. More than half of the mutations detected were classified as putative driver mutations of cancer, and virtually all affected genes recurrently mutated in MNs. Reflecting prior studies, the most commonly identified CH mutations affected epigenetic modifier genes DNMT3A and TET2, as well as DDR genes TP53, CHEK2, and PPM1D.
Correlation between CH and clinical characteristics was performed for 10,207 patients; 61 percent had exposure to cancer therapy prior to CH analysis, and 39 percent were treatment naïve. The authors found that CH was associated with increasing age (OR, 1.8; p≤10-6) and cell extrinsic factors such as smoking exposure (OR, 1.1; p=4.1 × 10-3) and previous exposure to cancer therapy (OR, 1.2; p=4.2 × 10-6). Although splicing factor mutations were less commonly detected, they were more strongly correlated with older age. TP53 (OR, 2.7; q=9.0 × 104), PPM1D (OR, 3.6; q=1.2 × 10-5), and CHEK2 (OR=4.6, q≤106) mutations were significantly associated with previous exposure to cancer therapy, an association not seen for mutations in epigenetic modifiers or splicing factors. CH was most strongly associated with topoisomerase II inhibitors (OR, 1.3; p=0.01) and platinum drugs (OR, 1.2; p=0.01) — carboplatin specifically (OR, 1.3; p=0.002) — for the latter class. Consistent with these observations, carboplatin is particularly associated with risk of tMN.14 A dose-response relationship was observed between cancer therapy and the prevalence of CH; a higher cumulative exposure to external beam radiation and platinum chemotherapies was positively associated with the presence of CH. Specifically for TP53 mutations, significant associations were found in patients with exposure to platinum, taxanes, and radiation therapy.
Serial analysis in 525 patients allowed interrogation of the clonal dynamics of CH after cancer therapy. Sixty-two percent of persons with a CH mutation at both time points had a stable allele frequency, whereas in 28 percent of cases, mutations increased in VAF, and 10 percent showed a decrease in clone size. In patients receiving radiation or cytotoxic therapy, there was a selective increase in DDR mutations; moreover, increasing exposure to either radiation or cytotoxic therapy led to a further increase in allele burdens of DDR mutations. This highlights the relationship between cancer therapy and TP53, PPM1D, and CHEK2 mutations; exposure to specific cytotoxic treatments and radiotherapy confers a competitive growth advantage in patients with cancer harboring a DDR mutant clone. TP53 mutations were among the strongest risk factors associated with subsequent progression to tMN in an analysis of almost 10,000 patients exposed to cancer therapies. Indeed, a further analysis of tMN patients detected TP53 mutations in 40 percent (n=14/35 patients), which were present in a pretransformation paired sample in 10 of 14 patients (median intersampling time was 24 months for these patients).
There are numerous important ramifications of these large cohort analyses of cancer and MDS/tMN patients. Monitoring for CH may have a predictive role in risk stratifying patients with cancer to identify those at increased risk of tMN. This would have the potential to influence therapeutic decisions made during their cancer treatment. Dr. Bolton and colleagues investigated how CH may be incorporated into a risk stratification model for tMN, including other relevant parameters such as age and blood counts using an established methodology (iCARE). They modelled the 10-year risk of MDS/AML in women with breast cancer, estimating the risk for those receiving and not receiving adjuvant chemotherapy. In patients with the highest risk of tMN (top 1%), the estimated increase in risk with the addition of adjuvant chemotherapy was greater than 9 percent, which would exceed the predicted survival benefit of adjuvant chemotherapy in many women with breast cancer. Furthermore, the findings of Dr. Bernard and colleagues suggest that identification of biallelic TP53 mutations could be used to further refine this risk stratification. Ultimately, single-cell analysis might be required to definitively identify subclonal biallelic TP53 mutations. Such an approach will require prospective validation before it is incorporated into routine clinical practice. It will also be important to develop clear recommendations to guide oncologists in the appropriate use of CH screening to enhance optimal therapeutic decision making.
References
Competing Interests
Dr. Mead indicated no relevant conflicts of interest.