Although most patients with acute myeloid leukemia (AML) achieve remission after intensive chemotherapy, relapse remains common.1 Recurrent genetic alterations are frequently detected by next-generation sequencing (NGS) in AML and have important therapeutic and prognostic implications.2 The ability to detect persistent mutations in patients with AML in remission after therapy (termed “molecular measurable residual disease” [MRD]) has enabled the identification of patients at high risk of relapse.1,3-5 However, not all mutations are created equal in terms of their clinical implications: While the persistence of certain genetic aberrations such as mutated NPM1 or a PML-RARA rearrangement has been shown to correlate with risk of relapse, mutations in the epigenetic regulator genes DNMT3A, TET2, and ASXL1 (so-called “DTA mutations”) do not seem to be associated with an adverse outcome. The latter are thought to reflect the persistence of preleukemic age-related clonal hematopoiesis of indeterminate potential (CHIP) rather than true residual AML.1 Distinguishing between these relatively innocuous CHIP mutations and true leukemic driver mutations remains uncertain, yet is of the utmost importance to determine risk of relapse for patients with AML in remission. In particular, polymerase chain reaction (PCR) detection of NPM1 mutation has proven to be a highly sensitive and specific method for identifying and tracking MRD in NPM1-mutated AML and correlates well with relapse risk.6
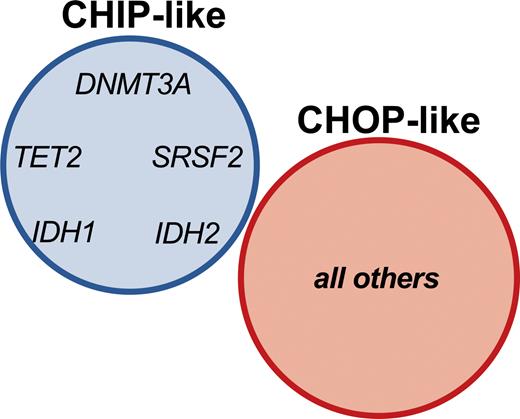
Dr. Luca Vincenzo Cappelli and colleagues sought to establish the significance of mutational profiles over time in 150 patients diagnosed with NPM1-mutated AML who achieved complete molecular remission (CMR; the absence of evidence of detectable NPM1 mutation by sensitive PCR assay). The authors found that 46 percent of patients in CMR harbored one or more mutations with a variant allele frequency of at least 1 percent. Mutations that tended to be present at diagnosis, CMR, and relapse included the expected DTA genes DNMT3A and TET2, but interestingly also included mutations in IDH1, IDH2, and SRSF2. In contrast, mutations in PTPN11, WT1, CEBP1, GATA2, and RUNX1 tended to be absent at diagnosis and were acquired at CMR or relapse. The authors proposed that the former “CHIP-like” mutations represent a pre-leukemic state onto which oncogenic driver mutations are acquired; this model was supported by clonal hierarchy analysis at diagnosis, which revealed that CHIP-like mutations generally showed a higher variant allele frequency relative to the NPM1 mutation. Patients harboring CHIP-like mutations at CMR showed superior overall survival and event-free survival relative to patients with other mutations. Importantly, the addition of IDH1, IDH2, and SRSF2 to the CHIP-like group showed a stronger predictive power relative to the DTA-only group, indicating that identification of these mutations during CMR does not imply a higher risk of relapse and may not warrant additional therapy. Conversely, other mutations that were identified as present during CMR were associated with high risk of relapse, and thus appeared to reflect true oncogenic drivers, termed “clonal hematopoiesis of oncogenic potential,” or “CHOP-like” mutations (Figure). Overall, these findings highlight the importance of interrogating bone marrow samples at the time of CMR not only to sensitively assess for an NPM1 mutation indicating MRD, but also by NGS to assess for CHOP-like mutations that may also have implications in predicting high likelihood of relapse.7 Despite clinical remission, patients with CHOP-like mutations may benefit from additional maintenance therapies such as azacytidine.8 The findings also underscore the genetic complexity of post-treatment AML, which frequently harbors clonal hematopoiesis with different implications depending on the specific mutation profile. Some post-AML clonal hematopoiesis states may be accompanied by abnormal immunophenotypes and morphologic dysplasia of background hematopoietic elements, which have as of yet uncertain clinical implications.9
Clonal hematopoiesis of indeterminate potential (CHIP) vs. clonal hematopoiesis of oncogenic potential (CHOP) mutations in NPM1-mutated acute myeloid leukemia. CHIP-like mutations are defined as the DTA mutations DNMT3A and TET2 in addition to IDH1, IDH2, and SRSF2 detected at complete molecular remission (CMR). CHOP-like mutations include all other mutations detected at CMR. From Cappelli LV, Meggendorfer M, Baer C, et al. Indeterminate and oncogenic potential: CHIP vs CHOP mutations in AML with NPM1 alteration. Leukemia 2021.
Clonal hematopoiesis of indeterminate potential (CHIP) vs. clonal hematopoiesis of oncogenic potential (CHOP) mutations in NPM1-mutated acute myeloid leukemia. CHIP-like mutations are defined as the DTA mutations DNMT3A and TET2 in addition to IDH1, IDH2, and SRSF2 detected at complete molecular remission (CMR). CHOP-like mutations include all other mutations detected at CMR. From Cappelli LV, Meggendorfer M, Baer C, et al. Indeterminate and oncogenic potential: CHIP vs CHOP mutations in AML with NPM1 alteration. Leukemia 2021.
In Brief
The work of Dr. Cappelli and colleagues has refined our understanding of the influence of the clonal background on treated NPM1-mutated AML. The authors expand the category of DTA mutations in this context to also include IDH1, IDH2, and SRSF2, and confirm that the persistence of these CHIP-like mutations at CMR does not appear to negatively impact outcome in NPM1-mutated AML. In contrast, identification of other mutations at CMR defines a group of patients with adverse prognosis and higher risk of relapse who may benefit from additional therapy. Clinical trials focused on patients harboring these CHOP-like mutations at CMR are needed to better establish appropriate treatment strategies to eradicate low levels of disease that are harbingers of relapse.
Competing Interests
Dr. Fitzpatrick and Dr. Hasserjian indicated no relevant conflicts of interest.