Patients with diffuse large B-cell lymphoma (DLBCL) display variable clinical outcomes despite generally uniform treatment. A major goal of improving outcomes of those with DLBCL is to use individual patient characteristics to tailor treatment. While the International Prognostic Index is useful in predicting overall survival based on clinical factors, it does not take into account biologic features.1 For approximately 25 years, it has been recognized that DLBCL arises from at least two principal types of B cells, termed “cell of origin.” When treated with standard chemoimmunotherapy, DLBCL arising from germinal center B-cells tends to have more favorable clinical outcomes than DLBCL arising from activated B-cells (ABC).2
Large-scale, next-generation sequencing (NGS) inquiries identified recurring driver somatic mutations responsible for DLBCL pathogenesis.3 More recently, integrative multiomics approaches have combined NGS and assessment of additional molecular abnormalities, including chromosomal structural variants (SVs) such as translocations, insertions, deletions, and copy number alterations (CNAs), to categorize DLBCL into additional subgroups or clusters.4,5
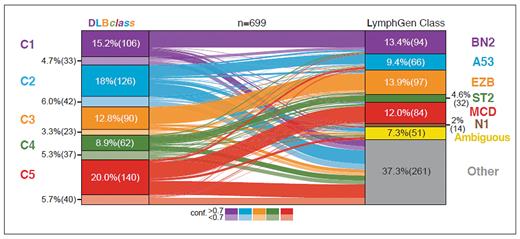
The most widely available system for advanced classification of DLBCL is the LymphGen algorithm (llmpp.nih.gov/lymphgen/index.php), which classifies DLBCL into six different subgroups (Table). As first described by Roland Schmitz, PhD, and then modified by George Wright, MD, both working with colleagues in the laboratory of Louis M. Staudt, MD, PhD, these subgroups are highly analogous to clusters independently described by Bjoern Chapuy, MD, PhD, and colleagues working with Margaret A. Shipp, MD, in 2018.4-6 For instance, patients with DLBCL whose tumors belong to the MCD LymphGen subtype have malignant cells that are dependent on B-cell receptor signaling. These typically have an ABC gene expression profile and frequently harbor BCL2 copy gains and the MYD88 L265P mutation, as well as mutations in CD79b. MCD tumors are highly similar to C5 tumors in the system described by Dr. Chapuy and colleagues. Targeted inhibition of these pathways with Bruton’s tyrosine kinase inhibitors is highly likely to be effective in patients harboring this type of lymphoma.7 As shown in the figure below, the other LymphGen subtypes (BN2, A53, EZB, and ST2) are analogous to individual clusters (C1, C2, C3, and C4, respectively).
Comparison of DLBclass cluster assignments and LymphGen subsets
Comparison of DLBclass cluster assignments and LymphGen subsets
Comparison of diffuse large B-cell lymphoma subclassification systems
| Cell of origin . | Risk level . | Method of DLBCL classification . | Recurring molecular abnormalities . | |
|---|---|---|---|---|
| DLBclass . | LymphGen6 . | |||
| Germinal center | Low | Cluster 4 | ST2 | Histone mutations JAK-STAT and PI3K signaling NF-κB mutations |
| High | Cluster 3 | EZB | BCL2 translocations EZH2 activating mutations PI3K signaling | |
| Activated B-cell | Low | Cluster 1 | BN2 | Immune evasion profile NOTCH2/NF-κB alterations BCL6 translocations MYD88 non-L265P |
| High | Cluster 5 | MCD | CD79b MYD88 L265P 18q gains BCL2 expression | |
| N1 | NOTCH1 mutations | |||
| Both | High | Cluster 2 | A53 | Inactivation of p53 ± CDKN2A Aneuploidy |
| Cell of origin . | Risk level . | Method of DLBCL classification . | Recurring molecular abnormalities . | |
|---|---|---|---|---|
| DLBclass . | LymphGen6 . | |||
| Germinal center | Low | Cluster 4 | ST2 | Histone mutations JAK-STAT and PI3K signaling NF-κB mutations |
| High | Cluster 3 | EZB | BCL2 translocations EZH2 activating mutations PI3K signaling | |
| Activated B-cell | Low | Cluster 1 | BN2 | Immune evasion profile NOTCH2/NF-κB alterations BCL6 translocations MYD88 non-L265P |
| High | Cluster 5 | MCD | CD79b MYD88 L265P 18q gains BCL2 expression | |
| N1 | NOTCH1 mutations | |||
| Both | High | Cluster 2 | A53 | Inactivation of p53 ± CDKN2A Aneuploidy |
Integrated multiomics methods include simultaneous integration of data from next-generation sequencing and gene expression profiling, as well as assessment of chromosome structural variants and copy number alterations. Abbreviation: DLBCL, diffuse large B-cell lymphoma.
The knowledge of DLBCL subtype could be used to tailor treatment by the addition of specific pathway inhibitors to standard therapy in frontline clinical trials, as well as potentially in the relapsed setting. Genomic subclassification of tumors from formalin-fixed, paraffin-embedded tissue biopsies requires a complex and potentially costly diagnostic test. Therefore, it is of great interest to have the maximal number of cases classified as possible. In the current iteration, LymphGen assigns a single subgroup for approximately 60% of patient’s tumor samples, leaving uncertainty about the best therapeutic candidates for nearly half of patients.
In a 2024 article in Blood, Dr. Chapuy and colleagues working in the laboratory of Gad Getz, PhD, present an updated classification for DLBCL, termed DLBclass, that aims to classify all DLBCL tumors. Like previous work, the classification system is based on somatic mutations (as well as SV and CNA assessment) and assigns DLBCLs into one of five genetic clusters, C1 to C5. However, as an improvement on prior work, the investigators leveraged advanced machine learning tools and merged data from the LymphGen training set with their own to arrive at DLBclass, which can classify nearly all cases to one of these five clusters with varying degrees of confidence. Cases that have a high degree of similarity to the core group are scored with high confidence, while those with molecular features that have fewer core features are assigned a lower confidence score. While there are likely to be some cases that do not have significant similarity to each cluster, the new system offers the broadest classification schema and is publicly available at github.com/getzlab/DLBCL-Classifier.
In Brief
What are the implications of DLBclass? As advanced molecular assays become more widely available for real-time testing, future clinical trials of novel agents for DLBCL are likely to integrate such approaches. Additionally, as there has been an explosion of novel therapeutic agents for patients with DLBCL, including bispecific T-cell engaging therapies, antibody drug conjugates, and novel cellular therapies, it will be important for prospective trials to have a plan for assessing molecular subtypes of archival tissues from patients obtained at the time of study entry. In the current era, DLBclass offers the system capable of classifying the greatest number of patients to gain insight into which intervention is most likely to be beneficial. With the caveat that the biologically heterogeneity of DLBCL will always impose inherent limits on the degree to which any classification system can categorize patients into discrete groups, the end result of DLBclass is a welcome and important tool that should help bring the field closer to the goal of testing personalized treatment for all patients.
Disclosure Statement
Dr. Hill indicated no relevant conflicts of interest.